Application of m6A and TME in Predicting the Prognosis and Treatment of Clear Cell Renal Cell Carcinoma
- PMID: 35281520
- PMCID: PMC8916893
- DOI: 10.1155/2022/2910491
Application of m6A and TME in Predicting the Prognosis and Treatment of Clear Cell Renal Cell Carcinoma
Abstract
Background: Previous studies have shown that RNA N6-methyladenosine (m6A) plays an important role in the construction of the tumor microenvironment (TME). However, how m6A plays a role in the TME of clear cell renal cell carcinoma remains unclear.
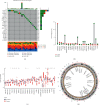
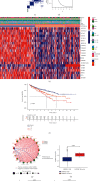
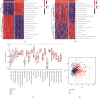
Methods: Based on 23 m6A modulators, we applied consensus cluster analysis to explore the different m6A modification profiles of ccRCC. The CIBERSORT method was employed to reveal the correlation between TME immune cell infiltration and different m6A modification patterns. A m6A score was constructed using a principal component analysis algorithm to assess and quantify the m6A modification patterns of individual tumors.
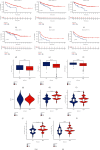
Results: Three distinct m6A modification patterns of ccRCC were identified. The characteristics of TME cell infiltration in these three patterns were consistent with immune rejection phenotype, immune inflammation phenotype, and immune desert phenotype. In particular, when m6A scores were high, TME was characterized by immune cell infiltration and patient survival was higher (p < 0.05). When m6A scores were low, TME was characterized by immunosuppression and patient survival was lower (p < 0.05). The immunotherapy cohort confirmed that patients with higher m6A scores had significant therapeutic advantages and clinical benefits.
Conclusions: The m6A modification plays an important role in the formation of TME. The m6A scoring system allows the identification of m6A modification patterns in individual tumors, discriminates the immune infiltrative features of TME, and provides more effective prognostic indicators and treatment strategies for immunotherapy.
Copyright © 2022 Dongchen Pei et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



Similar articles
-
m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer.Mol Cancer. 2020 Mar 12;19(1):53. doi: 10.1186/s12943-020-01170-0. Mol Cancer. 2020. PMID: 32164750 Free PMC article.
-
m6A regulator-based methylation modification patterns characterized by distinct tumor microenvironment immune profiles in colon cancer.Theranostics. 2021 Jan 1;11(5):2201-2217. doi: 10.7150/thno.52717. eCollection 2021. Theranostics. 2021. PMID: 33500720 Free PMC article.
-
Integrative 5-Methylcytosine Modification Immunologically Reprograms Tumor Microenvironment Characterizations and Phenotypes of Clear Cell Renal Cell Carcinoma.Front Cell Dev Biol. 2021 Dec 8;9:772436. doi: 10.3389/fcell.2021.772436. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34957104 Free PMC article.
-
N6-Methyladenosine RNA Modification: An Emerging Immunotherapeutic Approach to Turning Up Cold Tumors.Front Cell Dev Biol. 2021 Sep 20;9:736298. doi: 10.3389/fcell.2021.736298. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34616742 Free PMC article. Review.
-
Progression of m6A in the tumor microenvironment: hypoxia, immune and metabolic reprogramming.Cell Death Discov. 2024 Jul 20;10(1):331. doi: 10.1038/s41420-024-02092-2. Cell Death Discov. 2024. PMID: 39033180 Free PMC article. Review.
Cited by
-
RNA N6-methyladenosine modifications in urological cancers: from mechanism to application.Nat Rev Urol. 2024 Aug;21(8):460-476. doi: 10.1038/s41585-023-00851-x. Epub 2024 Feb 12. Nat Rev Urol. 2024. PMID: 38347160 Review.
References
LinkOut - more resources
Full Text Sources

