Comparative N-Glycoproteomics Analysis of Clinical Samples Via Different Mass Spectrometry Dissociation Methods
- PMID: 35281567
- PMCID: PMC8907888
- DOI: 10.3389/fchem.2022.839470
Comparative N-Glycoproteomics Analysis of Clinical Samples Via Different Mass Spectrometry Dissociation Methods
Abstract
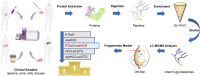
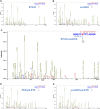
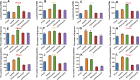
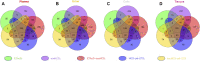
Site-specific N-glycosylation characterization requires intact N-glycopeptide analysis based on suitable tandem mass spectrometry (MS/MS) method. Electron-transfer/higher-energy collisional dissociation (EThcD), stepped collision energy/higher-energy collisional dissociation (sceHCD), higher-energy collisional dissociation-product-dependent electron-transfer dissociation (HCD-pd-ETD), and a hybrid mass spectrometry fragmentation method EThcD-sceHCD have emerged as valuable approaches for glycoprotein analysis. However, each of them incurs some compromise, necessitating the systematic performance comparisons when applied to the analysis of complex clinical samples (e.g., plasma, urine, cells, and tissues). Herein, we compared the performance of EThcD-sceHCD with those previous approaches (EThcD, sceHCD, HCD-pd-ETD, and sceHCD-pd-ETD) in the intact N-glycopeptide analysis, and determined its applicability for clinical N-glycoproteomic study. The intact N-glycopeptides of distinct samples, namely, plasma from prostate cancer (PCa) patients, urine from immunoglobulin A nephropathy (IgAN) patients, human hepatocarcinoma cell line (HepG2), and thyroid tissues from thyroid cancer (TC) patients were analyzed by these methods. We found that EThcD-sceHCD outperformed other methods in the balance of depth and accuracy of intact N-glycopeptide identification, and sceHCD and EThcD-sceHCD have good complementarity. EThcD-sceHCD holds great potential for biomarker discovery from clinical samples.
Keywords: N-glycosylation; clinical sample; electron-transfer/higher-energy collisional dissociation (EThcD)-stepped collision energy/higher-energy collisional dissociation (sceHCD); glycoproteomics; mass spectrometry.
Copyright © 2022 Zeng, Zheng, Su, Cheng, Mao, Zhong, Liu, Chen, Zhao, Lin, Liu, Li, Yang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Comprehensive Plasma N-Glycoproteome Profiling Based on EThcD-sceHCD-MS/MS.Front Chem. 2022 Jun 20;10:920009. doi: 10.3389/fchem.2022.920009. eCollection 2022. Front Chem. 2022. PMID: 35795219 Free PMC article.
-
Optimal Dissociation Methods Differ for N- and O-Glycopeptides.J Proteome Res. 2020 Aug 7;19(8):3286-3301. doi: 10.1021/acs.jproteome.0c00218. Epub 2020 Jun 28. J Proteome Res. 2020. PMID: 32500713 Free PMC article.
-
Site-specific N-glycosylation characterization of micro monoclonal immunoglobulins based on EThcD-sceHCD-MS/MS.Front Immunol. 2022 Sep 15;13:1013990. doi: 10.3389/fimmu.2022.1013990. eCollection 2022. Front Immunol. 2022. PMID: 36189210 Free PMC article.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
Cited by
-
Detection of N‑glycoprotein associated with IgA nephropathy in urine as a potential diagnostic biomarker using glycosylated proteomic analysis.Exp Ther Med. 2023 Aug 23;26(4):478. doi: 10.3892/etm.2023.12177. eCollection 2023 Oct. Exp Ther Med. 2023. PMID: 37753295 Free PMC article.
-
Uncovering the fragmentation and separation characteristics of sophorolipid biosurfactants with LC-MS-ESI.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae035. doi: 10.1093/jimb/kuae035. J Ind Microbiol Biotechnol. 2024. PMID: 39327028 Free PMC article.
-
Robust Glycoproteomics Platform Reveals a Tetra-Antennary Site-Specific Glycan Capping with Sialyl-Lewis Antigen for Early Detection of Gastric Cancer.Adv Sci (Weinh). 2024 Mar;11(9):e2306955. doi: 10.1002/advs.202306955. Epub 2023 Dec 12. Adv Sci (Weinh). 2024. PMID: 38084450 Free PMC article.
-
Comprehensive Plasma N-Glycoproteome Profiling Based on EThcD-sceHCD-MS/MS.Front Chem. 2022 Jun 20;10:920009. doi: 10.3389/fchem.2022.920009. eCollection 2022. Front Chem. 2022. PMID: 35795219 Free PMC article.
-
Quantitative N-glycoproteomic analysis reveals glycosylation signatures of plasma immunoglobulin G in systemic sclerosis.Front Immunol. 2025 Feb 7;16:1531191. doi: 10.3389/fimmu.2025.1531191. eCollection 2025. Front Immunol. 2025. PMID: 39991159 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

