Site-specific DNA functionalization through the tetrazene-forming reaction in ionic liquids
- PMID: 35282632
- PMCID: PMC8826848
- DOI: 10.1039/d1sc05204g
Site-specific DNA functionalization through the tetrazene-forming reaction in ionic liquids
Erratum in
-
Correction: Site-specific DNA functionalization through the tetrazene-forming reaction in ionic liquids.Chem Sci. 2022 May 30;13(22):6749-6751. doi: 10.1039/d2sc90102a. eCollection 2022 Jun 7. Chem Sci. 2022. PMID: 35756508 Free PMC article.
Abstract
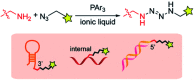
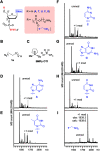
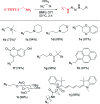
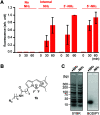
Development of multiple chemical tools for deoxyribonucleic acid (DNA) labeling has facilitated wide use of their functionalized conjugates, but significant practical and methodological challenges remain to achievement of site-specific chemical modification of the biomacromolecule. As covalent labeling processes are more challenging in aqueous solution, use of nonaqueous, biomolecule-compatible solvents such as an ionic liquid consisting of a salt with organic molecule architecture, could be remarkably helpful in this connection. Herein, we demonstrate site-specific chemical modification of unprotected DNAs through a tetrazene-forming amine-azide coupling reaction using an ionic liquid. This ionic liquid-enhanced reaction process has good functional group tolerance and precise chemoselectivity, and enables incorporation of various useful functionalities such as biotin, cholesterol, and fluorophores. A site-specifically labeled oligonucleotide, or aptamer interacting with a growth factor receptor (Her2) was successfully used in the fluorescence imaging of breast cancer cell lines. The non-traditional medium-promoted labeling strategy described here provides an alternative design paradigm for future development of chemical tools for applications involving DNA functionalization.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare the following competing financial interest(s): the authors have filed a patent application on the ionic-liquid-based tetrazene-forming bioconjugation method.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

