NMR methods for exploring 'dark' states in ligand binding and protein-protein interactions
- PMID: 35282867
- PMCID: PMC8921508
- DOI: 10.1016/j.pnmrs.2021.10.001
NMR methods for exploring 'dark' states in ligand binding and protein-protein interactions
Abstract
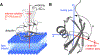
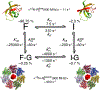
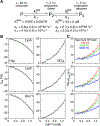
A survey, primarily based on work in the authors' laboratory during the last 10 years, is provided of recent developments in NMR studies of exchange processes involving protein-ligand and protein-protein interactions. We start with a brief overview of the theoretical background of Dark state Exchange Saturation Transfer (DEST) and lifetime line-broadening (ΔR2) NMR methodology. Some limitations of the DEST/ΔR2 methodology in applications to molecular systems with intermediate molecular weights are discussed, along with the means of overcoming these limitations with the help of closely related exchange NMR techniques, such as the measurements of Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion, exchange-induced chemical shifts or rapidly-relaxing components of relaxation decays. Some theoretical underpinnings of the quantitative description of global dynamics of proteins on the surface of very high molecular weight particles (nanoparticles) are discussed. Subsequently, several applications of DEST/ΔR2 methodology are described from a methodological perspective with an emphasis on providing examples of how kinetic and relaxation parameters for exchanging systems can be reliably extracted from NMR data for each particular model of exchange. Among exchanging systems that are not associated with high molecular weight species, we describe several exchange NMR-based studies that focus on kinetic modelling of transient pre-nucleation oligomerization of huntingtin peptides that precedes aggregation and fibril formation.
Keywords: Dark state exchange saturation transfer (DEST); Exchange-induced chemical shifts; Fast component of relaxation decay; Lifetime line-broadening; Relaxation dispersion.
Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

















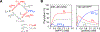



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

