TAILS Identifies Candidate Substrates and Biomarkers of ADAMTS7, a Therapeutic Protease Target in Coronary Artery Disease
- PMID: 35283288
- PMCID: PMC9035411
- DOI: 10.1016/j.mcpro.2022.100223
TAILS Identifies Candidate Substrates and Biomarkers of ADAMTS7, a Therapeutic Protease Target in Coronary Artery Disease
Abstract
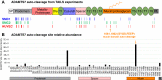
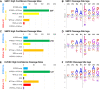
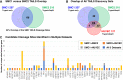
Loss-of-function mutations in the secreted enzyme ADAMTS7 (a disintegrin and metalloproteinase with thrombospondin motifs 7) are associated with protection for coronary artery disease. ADAMTS7 catalytic inhibition has been proposed as a therapeutic strategy for treating coronary artery disease; however, the lack of an endogenous substrate has hindered the development of activity-based biomarkers. To identify ADAMTS7 extracellular substrates and their cleavage sites relevant to vascular disease, we used TAILS (terminal amine isotopic labeling of substrates), a method for identifying protease-generated neo-N termini. We compared the secreted proteome of vascular smooth muscle and endothelial cells expressing either full-length mouse ADAMTS7 WT, catalytic mutant ADAMTS7 E373Q, or a control luciferase adenovirus. Significantly enriched N-terminal cleavage sites in ADAMTS7 WT samples were compared to the negative control conditions and filtered for stringency, resulting in catalogs of high confidence candidate ADAMTS7 cleavage sites from our three independent TAILS experiments. Within the overlap of these discovery sets, we identified 24 unique cleavage sites from 16 protein substrates, including cleavage sites in EFEMP1 (EGF-containing fibulin-like extracellular matrix protein 1/Fibulin-3). The ADAMTS7 TAILS preference for EFEMP1 cleavage at the amino acids 123.124 over the adjacent 124.125 site was validated using both endogenous EFEMP1 and purified EFEMP1 in a binary in vitro cleavage assay. Collectively, our TAILS discovery experiments have uncovered hundreds of potential substrates and cleavage sites to explore disease-related biological substrates and facilitate activity-based ADAMTS7 biomarker development.
Keywords: ADAMTS; ADAMTS7; Biomarkers; Coronary artery disease; Degradomics; EFEMP1; Mass spectrometry; Protease; Substrate identification; TAILS proteomics.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest B. T. M., N. H. E., and Y. X. are named inventors on patent applications relating to ADAMTS7 assays and compounds. B. T. M., A. A., and N. H. E are named inventors on a patent application relating to ADAMTS7 biomarkers. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures



References
-
- Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R., Horne B.D., Stewart A.F., Assimes T.L., Wild P.S., Allayee H., et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet. 2011;377:383–392. - PMC - PubMed
-
- Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., Absher D., Aherrahrou Z., Allayee H., Altshuler D., Anand S.S., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. - PMC - PubMed
-
- Coronary Artery Disease Genetics, C. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. - PubMed
-
- Pu X., Xiao Q., Kiechl S., Chan K., Ng F.L., Gor S., Poston R.N., Fang C., Patel A., Senver E.C., Shaw-Hawkins S., Willeit J., Liu C., Zhu J., Tucker A.T., et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am. J. Hum. Genet. 2013;92:366–374. - PMC - PubMed
-
- Pereira A., Palma Dos Reis R., Rodrigues R., Sousa A.C., Gomes S., Borges S., Ornelas I., Freitas A.I., Guerra G., Henriques E., Rodrigues M., Freitas S., Freitas C., Brehm A., Pereira D., et al. Association of ADAMTS7 gene polymorphism with cardiovascular survival in coronary artery disease. Physiol. Genomics. 2016;48:810–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

