Preventing autosomal-dominant hearing loss in Bth mice with CRISPR/CasRx-based RNA editing
- PMID: 35283480
- PMCID: PMC8918553
- DOI: 10.1038/s41392-022-00893-4
Preventing autosomal-dominant hearing loss in Bth mice with CRISPR/CasRx-based RNA editing
Abstract
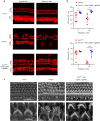
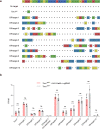
CRISPR/RfxCas13d (CasRx) editing system can specifically and precisely cleave single-strand RNAs, which is a promising treatment for various disorders by downregulation of related gene expression. Here, we tested this RNA-editing approach on Beethoven (Bth) mice, an animal model for human DFNA36 due to a point mutation in Tmc1. We first screened 30 sgRNAs in cell cultures and found that CasRx with sgRNA3 reduced the Tmc1Bth transcript by 90.8%, and the Tmc1 wild type transcript (Tmc1+) by 44.3%. We then injected a newly developed AAV vector (AAV-PHP.eB) based CasRx into the inner ears of neonatal Bth mice, and we found that Tmc1Bth was reduced by 70.2% in 2 weeks with few off-target effects in the whole transcriptome. Consistently, we found improved hair cell survival, rescued hair bundle degeneration, and reduced mechanoelectrical transduction current. Importantly, the hearing performance, measured in both ABR and DPOAE thresholds, was improved significantly in all ages over 8 weeks. We, therefore, have validated the CRISPR/CasRx-based RNA editing strategy in treating autosomal-dominant hearing loss, paving way for its further application in many other hereditary diseases in hearing and beyond.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- World Health Organization. Deafness and Hearing Loss. http://www.who.int/mediacentre/factsheets/fs300/en/ (2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

