Virulence genes and antimicrobial resistance pattern in Proteus mirabilis strains isolated from patients attended with urinary infections to Tertiary Hospitals, in Iran
- PMID: 35283944
- PMCID: PMC8889823
- DOI: 10.4314/ahs.v21i4.22
Virulence genes and antimicrobial resistance pattern in Proteus mirabilis strains isolated from patients attended with urinary infections to Tertiary Hospitals, in Iran
Abstract
Background: Proteus mirabilis is a frequent reason for catheter-associated urinary tract infections (UTIs). The aim of this study was to identify virulence genes and antimicrobial resistance patterns in P. mirabilis strains isolated from patients who attended a tertiary hospital in Iran.
Methods: In this study, 100 P. mirabilis strains from urine samples were isolated. These isolated strains were identified by biochemical and PCR-based tests, and their antibiotic resistance was profiled through a standard procedure using 14 antibiotics. PCR assays were used to detect virulence-related genes in P. mirabilis strains. The biofilm formation of each P. mirabilis strain was examined.
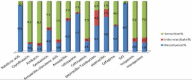
Results: Of the 100 P. mirabilis isolates, 16 (16%) were multidrug-resistant. High resistance was observed against cotrimoxazole (97%), nalidixic acid (93%), cefotaxime (77%), and amoxicillin (62%). Sixty of the 100 isolates showed resistance against extended-spectrum cephalosporins. The prevalence rates of the genes related to the virulence factors in this study were mrpH (100%), ucaA (91%), hpmA (94%), zapA (95%), ptaA (100%), ureG (100%), pmfA (100%), fliC (97%), and mrpA (90%) using PCR method. Strong biofilm formation was observed in 20% (5/25) of the strains isolated from non-catheterized samples and 80% (20/25) of strains isolated from catheterized samples.
Conclusions: Resistance to antibiotics and the prevalence of pathogenicity genes are high in Proteus mirabilis strains iolated from UTIs.
Keywords: Antibiotic resistance; Proteus mirabilis; biofilm; virulence factors.
© 2021 Tabatabaei A et al.
Figures
References
-
- Getliffe K, Newton T. Catheter-associated urinary tract infection in primary and community health care. Age and Ageing. 2006;35(5):477–481. - PubMed
-
- Prywer J, Olszynski M. Bacterially induced formation of infectious urinary stones: recent developments and future challenges. Current Medicinal Chemistry. 2017;24(3):292–311. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical