Design of SARS-CoV-2 Variant-Specific PCR Assays Considering Regional and Temporal Characteristics
- PMID: 35285246
- PMCID: PMC9004361
- DOI: 10.1128/aem.02289-21
Design of SARS-CoV-2 Variant-Specific PCR Assays Considering Regional and Temporal Characteristics
Abstract
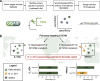
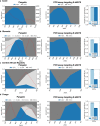
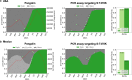
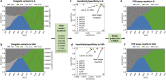
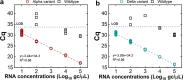
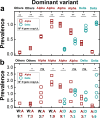
Monitoring the prevalence of SARS-CoV-2 variants is necessary to make informed public health decisions during the COVID-19 pandemic. PCR assays have received global attention, facilitating a rapid understanding of variant dynamics because they are more accessible and scalable than genome sequencing. However, as PCR assays target only a few mutations, their accuracy could be reduced when these mutations are not exclusive to the target variants. Here we introduce PRIMES, an algorithm that evaluates the sensitivity and specificity of SARS-CoV-2 variant-specific PCR assays across different geographical regions by incorporating sequences deposited in the GISAID database. Using PRIMES, we determined that the accuracy of several PCR assays decreased when applied beyond the geographic scope of the study in which the assays were developed. Subsequently, we used this tool to design Alpha and Delta variant-specific PCR assays for samples from Illinois, USA. In silico analysis using PRIMES determined the sensitivity/specificity to be 0.99/0.99 for the Alpha variant-specific PCR assay and 0.98/1.00 for the Delta variant-specific PCR assay in Illinois, respectively. We applied these two variant-specific PCR assays to six local sewage samples and determined the dominant SARS-CoV-2 variant of either the wild type, the Alpha variant, or the Delta variant. Using next-generation sequencing (NGS) of the spike (S) gene amplicons of the Delta variant-dominant samples, we found six mutations exclusive to the Delta variant (S:T19R, S:Δ156/157, S:L452R, S:T478K, S:P681R, and S:D950N). The consistency between the variant-specific PCR assays and the NGS results supports the applicability of PRIMES. IMPORTANCE Monitoring the introduction and prevalence of variants of concern (VOCs) and variants of interest (VOIs) in a community can help the local authorities make informed public health decisions. PCR assays can be designed to keep track of SARS-CoV-2 variants by measuring unique mutation markers that are exclusive to the target variants. However, the mutation markers may not be exclusive to the target variants because of regional and temporal differences in variant dynamics. We introduce PRIMES, an algorithm that enables the design of reliable PCR assays for variant detection. Because PCR is more accessible, scalable, and robust for sewage samples than sequencing technology, our findings will contribute to improving global SARS-CoV-2 variant surveillance.
Keywords: PCR assays; PRIMES; SARS-CoV-2 variants; in silico analysis; wastewater-based epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance.Microbiol Spectr. 2022 Feb 23;10(1):e0251321. doi: 10.1128/spectrum.02513-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196812 Free PMC article.
-
Two-Period Study Results from a Large Italian Hospital Laboratory Attesting SARS-CoV-2 Variant PCR Assay Evolution.Microbiol Spectr. 2022 Dec 21;10(6):e0292222. doi: 10.1128/spectrum.02922-22. Epub 2022 Nov 21. Microbiol Spectr. 2022. PMID: 36409091 Free PMC article.
-
CRISPR-Cas12a-Based Detection for the Major SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2021 Dec 22;9(3):e0101721. doi: 10.1128/Spectrum.01017-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787487 Free PMC article.
-
Molecular biology of SARS-CoV-2 and techniques of diagnosis and surveillance.Adv Clin Chem. 2024;118:35-85. doi: 10.1016/bs.acc.2023.11.003. Epub 2023 Dec 14. Adv Clin Chem. 2024. PMID: 38280807 Review.
-
Looking for a needle in a haystack. SARS-CoV-2 variant characterization in sewage.Curr Opin Environ Sci Health. 2021 Dec;24:100308. doi: 10.1016/j.coesh.2021.100308. Epub 2021 Nov 6. Curr Opin Environ Sci Health. 2021. PMID: 34849439 Free PMC article. Review.
Cited by
-
Efficient Tracing of the SARS-CoV-2 Omicron Variants in Santa Barbara County Using a Rapid Quantitative Reverse Transcription PCR Assay.Diagnostics (Basel). 2022 Nov 15;12(11):2805. doi: 10.3390/diagnostics12112805. Diagnostics (Basel). 2022. PMID: 36428863 Free PMC article.
-
Outbreak report of SARS-CoV-2 infection by airborne transmission: Epidemiologic and molecular evidence.Biomedica. 2023 Mar 30;43(1):121-130. doi: 10.7705/biomedica.6695. Biomedica. 2023. PMID: 37167462 Free PMC article.
-
Nucleic acid testing of SARS-CoV-2: A review of current methods, challenges, and prospects.Front Microbiol. 2022 Dec 9;13:1074289. doi: 10.3389/fmicb.2022.1074289. eCollection 2022. Front Microbiol. 2022. PMID: 36569096 Free PMC article. Review.
-
Improved performance of nucleic acid-based assays for genetically diverse norovirus surveillance.Appl Environ Microbiol. 2023 Oct 31;89(10):e0033123. doi: 10.1128/aem.00331-23. Epub 2023 Oct 4. Appl Environ Microbiol. 2023. PMID: 37791775 Free PMC article.
-
Application of neighborhood-scale wastewater-based epidemiology in low COVID-19 incidence situations.Sci Total Environ. 2022 Dec 15;852:158448. doi: 10.1016/j.scitotenv.2022.158448. Epub 2022 Sep 2. Sci Total Environ. 2022. PMID: 36063927 Free PMC article.
References
-
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. - DOI - PMC - PubMed
-
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wang M, Warren JL, Weinberger DM, Arnold W, Omer SB. 2020. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol 38:1164–1167. 10.1038/s41587-020-0684-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

