Functionalised Cofactor Mimics for Interactome Discovery and Beyond
- PMID: 35286003
- PMCID: PMC9401033
- DOI: 10.1002/anie.202201136
Functionalised Cofactor Mimics for Interactome Discovery and Beyond
Abstract
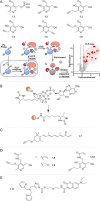
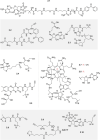
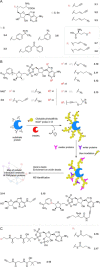
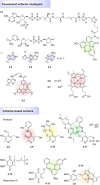
Cofactors are required for almost half of all enzyme reactions, but their functions and binding partners are not fully understood even after decades of research. Functionalised cofactor mimics that bind in place of the unmodified cofactor can provide answers, as well as expand the scope of cofactor activity. Through chemical proteomics approaches such as activity-based protein profiling, the interactome and localisation of the native cofactor in its physiological environment can be deciphered and previously uncharacterised proteins annotated. Furthermore, cofactors that supply functional groups to substrate biomolecules can be hijacked by mimics to site-specifically label targets and unravel the complex biology of post-translational protein modification. The diverse activity of cofactors has inspired the design of mimics for use as inhibitors, antibiotic therapeutics, and chemo- and biosensors, and cofactor conjugates have enabled the generation of novel enzymes and artificial DNAzymes.
Keywords: Chemical Probes; Cofactors; Photoaffinity Labelling; Protein Modifications; Proteomics.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Fischer J. D., Holliday G. L., Rahman S. A., Thornton J. M., J. Mol. Biol. 2010, 403, 803–824. - PubMed
-
- Richter M., Nat. Prod. Rep. 2013, 30, 1324–1345. - PubMed
-
- Katz E., Schlereth D. D., Schmidt H. L., Olsthoorn A. J. J., J. Electroanal. Chem. 1994, 368, 165–171.
-
- Xiao Y., Patolsky F., Katz E., Hainfeld J. F., Willner I., Science 2003, 299, 1877–1881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

