Multiple Introductions of Reassorted Highly Pathogenic Avian Influenza H5Nx Viruses Clade 2.3.4.4b Causing Outbreaks in Wild Birds and Poultry in The Netherlands, 2020-2021
- PMID: 35286149
- PMCID: PMC9045216
- DOI: 10.1128/spectrum.02499-21
Multiple Introductions of Reassorted Highly Pathogenic Avian Influenza H5Nx Viruses Clade 2.3.4.4b Causing Outbreaks in Wild Birds and Poultry in The Netherlands, 2020-2021
Abstract
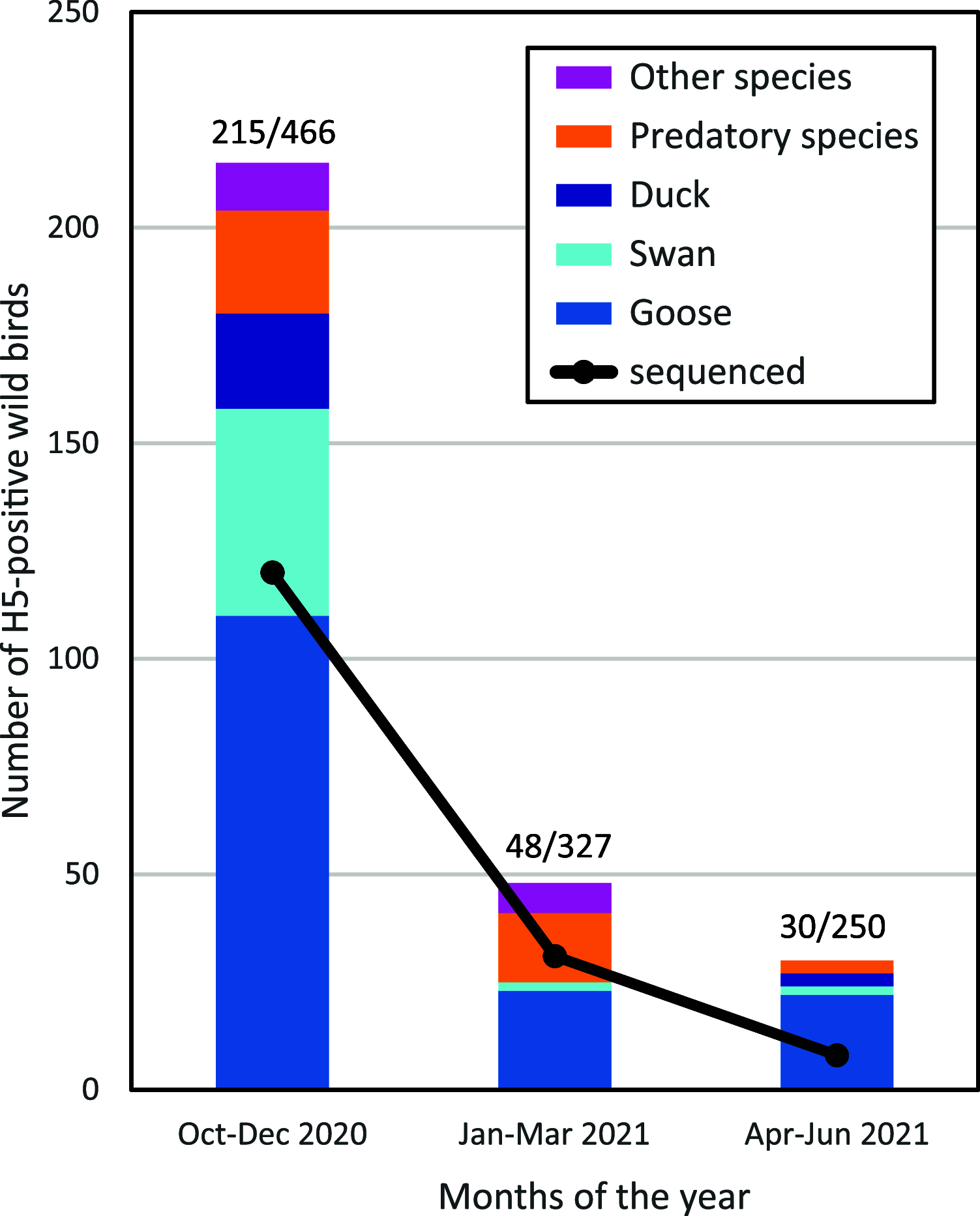
Highly pathogenic avian influenza (HPAI) viruses of subtype H5Nx caused outbreaks in poultry, captive birds, and wild birds in the Netherlands between October 2020 and June 2021. The full genome sequences of 143 viruses were analyzed. HPAI viruses were mainly of subtype H5N8, followed by H5N1, but also viruses of subtypes H5N3, H5N4, and H5N5 were detected. At least seven distinct genotypes were found, carrying closely related H5 segments belonging to clade 2.3.4.4b. Molecular clock analysis suggests that the reassortments of the NA gene segments likely occurred before the introduction of these viruses into the Netherlands. Genetic analysis suggested that multiple independent introductions of HPAI H5N8 viruses occurred in the Netherlands, likely followed by local spread resulting in at least two clusters of related viruses. The analysis provided evidence for independent introductions from wild birds at 10 poultry farms, whereas for two outbreaks transmission between farms could not be excluded. HPAI H5Nx viruses were detected in dead wild birds of 33 species, but mostly infected geese and swans were found. The pathogenicity of the H5N8 virus was determined for chickens and Pekin ducks, showing reduced mortality for ducks. This study provides more insight into the genetic diversity of HPAI H5Nx viruses generated by reassortment and evolution, and the spread of these viruses between wild birds and poultry. The fast and continuing evolution of H5 clade 2.3.4.4b may provide opportunities for these viruses to adapt to specific bird species, or possibly mammalian species and humans. IMPORTANCE Highly pathogenic avian influenza (HPAI) viruses are spread by migratory wild birds. Viruses causing outbreaks in wild birds and poultry in the Netherlands in 2020-2021 were genetically analyzed, which suggested that multiple virus incursions occurred. The outbreaks in poultry were likely caused by independent introductions from wild birds; only in one case virus spread between farms could not be excluded. Viruses of subtype H5N8 were mainly observed, but also other subtypes were detected that likely evolved by exchange of genetic information before these viruses were introduced into the Netherlands. Viruses were detected in many species of dead wild birds, but mostly in geese and swans. We showed that the H5N8 virus causes a higher mortality in chickens compared to ducks. This is consistent with the fact that not many wild ducks were found dead. This study provides more insight in the evolution and spread of HPAI viruses in wild birds and poultry.
Keywords: H5N8; HPAI; avian influenza virus; introduction routes; outbreak; reassortment; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen v.V, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J, World Health Organization International Avian Influenza Investigative Team. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 350:1179–1188. doi: 10.1056/NEJMoa040419. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

