METTL3 modulates m6A modification of CDC25B and promotes head and neck squamous cell carcinoma malignant progression
- PMID: 35287752
- PMCID: PMC8919647
- DOI: 10.1186/s40164-022-00256-3
METTL3 modulates m6A modification of CDC25B and promotes head and neck squamous cell carcinoma malignant progression
Abstract
Background: N6-methyladenosine (m6A) RNA methylation and its methyltransferase METTL3 have been widely reported to be involved in different cancers by regulating RNA metabolism and function. Here, we aimed to explore the biological function and clinical significance of m6A modification and METTL3 in head and neck squamous cell carcinoma (HNSCC).
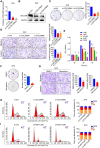
Methods: The prognostic value of METTL3 expression was evaluated using tissue microarray and immunohistochemical staining analyses in a human HNSCC cohort. The biological role and mechanism of METTL3 in HNSCC tumour growth, metastasis and angiogenesis were determined in vitro and in vivo.
Results: M6A levels and METTL3 expressions in HNSCC tissues were significantly increased compared with paired adjacent tissues. Meanwhile, METTL3 was an independent risk factor for the prognosis of HNSCC patients. Moreover, METTL3 overexpression promoted HNSCC cell proliferation, migration, invasion, and angiogenesis, while knockdown of METTL3 had an opposite effect in vivo and in vitro. Mechanistically, METTL3 enhanced the m6A modification of CDC25B mRNA, which maintained its stability and upregulated its expression, thereby activating G2/M phase of cell cycle and leading to HNSCC malignant progression.
Conclusions: METTL3 may be a potential prognostic biomarker and therapeutic target for HNSCC.
Keywords: CDC25B; Cell cycle; HNSCC; METTL3; m6A.
© 2022. The Author(s).
Conflict of interest statement
None.
Figures





References
-
- Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: a continuing challenge. Oncology. 2016;91(1):13–23. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Ca-a Cancer J Clin. 2019;69(1):7–34. - PubMed
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

