Defining A Global Map of Functional Group-based 3D Ligand-binding Motifs
- PMID: 35288344
- PMCID: PMC9881048
- DOI: 10.1016/j.gpb.2021.08.014
Defining A Global Map of Functional Group-based 3D Ligand-binding Motifs
Abstract
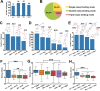
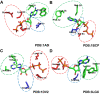
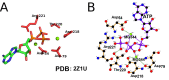
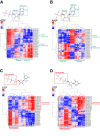
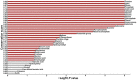
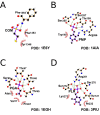
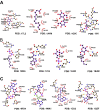
Uncovering conserved 3D protein-ligand binding patterns on the basis of functional groups (FGs) shared by a variety of small molecules can greatly expand our knowledge of protein-ligand interactions. Despite that conserved binding patterns for a few commonly used FGs have been reported in the literature, large-scale identification and evaluation of FG-based 3D binding motifs are still lacking. Here, we propose a computational method, Automatic FG-based Three-dimensional Motif Extractor (AFTME), for automatic mapping of 3D motifs to different FGs of a specific ligand. Applying our method to 233 naturally-occurring ligands, we define 481 FG-binding motifs that are highly conserved across different ligand-binding pockets. Systematic analysis further reveals four main classes of binding motifs corresponding to distinct sets of FGs. Combinations of FG-binding motifs facilitate the binding of proteins to a wide spectrum of ligands with various binding affinities. Finally, we show that our FG-motif map can be used to nominate FGs that potentially bind to specific drug targets, thus providing useful insights and guidance for rational design of small-molecule drugs.
Keywords: Binding motif; Computational method; Drug design; Functional group; Protein–ligand interaction.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





Similar articles
-
LibME-automatic extraction of 3D ligand-binding motifs for mechanistic analysis of protein-ligand recognition.FEBS Open Bio. 2016 Nov 30;6(12):1331-1340. doi: 10.1002/2211-5463.12150. eCollection 2016 Dec. FEBS Open Bio. 2016. PMID: 28255540 Free PMC article.
-
Structural motifs recurring in different folds recognize the same ligand fragments.BMC Bioinformatics. 2009 Jun 15;10:182. doi: 10.1186/1471-2105-10-182. BMC Bioinformatics. 2009. PMID: 19527512 Free PMC article.
-
Coagulase and Efb of Staphylococcus aureus Have a Common Fibrinogen Binding Motif.mBio. 2016 Jan 5;7(1):e01885-15. doi: 10.1128/mBio.01885-15. mBio. 2016. PMID: 26733070 Free PMC article.
-
Use of secondary structure element information in drug design: polypharmacology and conserved motifs in protein-ligand binding and protein-protein interfaces.Future Med Chem. 2011 Apr;3(6):699-708. doi: 10.4155/fmc.11.26. Future Med Chem. 2011. PMID: 21554076 Review.
-
Compound activity prediction using models of binding pockets or ligand properties in 3D.Curr Top Med Chem. 2012;12(17):1869-82. doi: 10.2174/156802612804547335. Curr Top Med Chem. 2012. PMID: 23116466 Free PMC article. Review.
References
-
- Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

