Lysosomal Ca2+-mediated TFEB activation modulates mitophagy and functional adaptation of pancreatic β-cells to metabolic stress
- PMID: 35288580
- PMCID: PMC8921223
- DOI: 10.1038/s41467-022-28874-9
Lysosomal Ca2+-mediated TFEB activation modulates mitophagy and functional adaptation of pancreatic β-cells to metabolic stress
Abstract
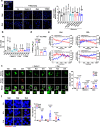
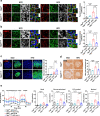
Although autophagy is critical for pancreatic β-cell function, the role and mechanism of mitophagy in β-cells are unclear. We studied the role of lysosomal Ca2+ in TFEB activation by mitochondrial or metabolic stress and that of TFEB-mediated mitophagy in β-cell function. Mitochondrial or metabolic stress induced mitophagy through lysosomal Ca2+ release, increased cytosolic Ca2+ and TFEB activation. Lysosomal Ca2+ replenishment by ER- > lysosome Ca2+ refilling was essential for mitophagy. β-cell-specific Tfeb knockout (TfebΔβ-cell) abrogated high-fat diet (HFD)-induced mitophagy, accompanied by increased ROS and reduced mitochondrial cytochrome c oxidase activity or O2 consumption. TfebΔβ-cell mice showed aggravation of HFD-induced glucose intolerance and impaired insulin release. Metabolic or mitochondrial stress induced TFEB-dependent expression of mitophagy receptors including Ndp52 and Optn, contributing to the increased mitophagy. These results suggest crucial roles of lysosomal Ca2+ release coupled with ER- > lysosome Ca2+ refilling and TFEB activation in mitophagy and maintenance of pancreatic β-cell function during metabolic stress.
© 2022. The Author(s).
Conflict of interest statement
M.-S.L. is the CEO of LysoTech, Inc. All authors declare no competing interests.
Figures




References
-
- Jung HS, et al. Loss of autophagy diminishes pancreatic b-cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

