Towards an efficient validation of dynamical whole-brain models
- PMID: 35288595
- PMCID: PMC8921267
- DOI: 10.1038/s41598-022-07860-7
Towards an efficient validation of dynamical whole-brain models
Abstract
Simulating the resting-state brain dynamics via mathematical whole-brain models requires an optimal selection of parameters, which determine the model's capability to replicate empirical data. Since the parameter optimization via a grid search (GS) becomes unfeasible for high-dimensional models, we evaluate several alternative approaches to maximize the correspondence between simulated and empirical functional connectivity. A dense GS serves as a benchmark to assess the performance of four optimization schemes: Nelder-Mead Algorithm (NMA), Particle Swarm Optimization (PSO), Covariance Matrix Adaptation Evolution Strategy (CMAES) and Bayesian Optimization (BO). To compare them, we employ an ensemble of coupled phase oscillators built upon individual empirical structural connectivity of 105 healthy subjects. We determine optimal model parameters from two- and three-dimensional parameter spaces and show that the overall fitting quality of the tested methods can compete with the GS. There are, however, marked differences in the required computational resources and stability properties, which we also investigate before proposing CMAES and BO as efficient alternatives to a high-dimensional GS. For the three-dimensional case, these methods generated similar results as the GS, but within less than 6% of the computation time. Our results contribute to an efficient validation of models for personalized simulations of brain dynamics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

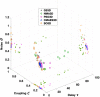


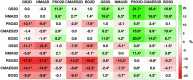






References
-
- van den Heuvel, M. P. & Hulshoff Pol, H. E. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol20, 519–534. 10.1016/j.euroneuro.2010.03.008 (2010). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

