Comprehensive biomarker profiles and chemometric filtering of urinary metabolomics for effective discrimination of prostate carcinoma from benign hyperplasia
- PMID: 35288652
- PMCID: PMC8921285
- DOI: 10.1038/s41598-022-08435-2
Comprehensive biomarker profiles and chemometric filtering of urinary metabolomics for effective discrimination of prostate carcinoma from benign hyperplasia
Abstract
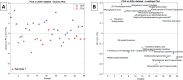
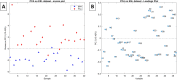
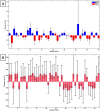
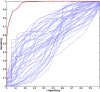
Prostate cancer (PCa) is the most commonly diagnosed cancer in male individuals, principally affecting men over 50 years old, and is the leading cause of cancer-related deaths. Actually, the measurement of prostate-specific antigen level in blood is affected by limited sensitivity and specificity and cannot discriminate PCa from benign prostatic hyperplasia patients (BPH). In the present paper, 20 urine samples from BPH patients and 20 from PCa patients were investigated to develop a metabolomics strategy useful to distinguish malignancy from benign hyperplasia. A UHPLC-HRMS untargeted approach was carried out to generate two large sets of candidate biomarkers. After mass spectrometric analysis, an innovative chemometric data treatment was employed involving PLS-DA classification with repeated double cross-validation and permutation test to provide a rigorously validated PLS-DA model. Simultaneously, this chemometric approach filtered out the most effective biomarkers and optimized their relative weights to yield the highest classification efficiency. An unprecedented portfolio of prostate carcinoma biomarkers was tentatively identified including 22 and 47 alleged candidates from positive and negative ion electrospray (ESI+ and ESI-) datasets. The PLS-DA model based on the 22 ESI+ biomarkers provided a sensitivity of 95 ± 1% and a specificity of 83 ± 3%, while that from the 47 ESI- biomarkers yielded an 88 ± 3% sensitivity and a 91 ± 2% specificity. Many alleged biomarkers were annotated, belonging to the classes of carnitine and glutamine metabolites, C21 steroids, amino acids, acetylcholine, carboxyethyl-hydroxychroman, and dihydro(iso)ferulic acid.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

