Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy
- PMID: 35288695
- PMCID: PMC9434490
- DOI: 10.1038/s41591-022-01702-9
Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy
Erratum in
-
Author Correction: Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy.Nat Med. 2023 Nov;29(11):2954. doi: 10.1038/s41591-022-02069-7. Nat Med. 2023. PMID: 36253610 Free PMC article. No abstract available.
Abstract
Anti-CD19 chimeric antigen receptor (CAR) T cell therapy has led to unprecedented responses in patients with high-risk hematologic malignancies. However, up to 60% of patients still experience disease relapse and up to 80% of patients experience CAR-mediated toxicities, such as cytokine release syndrome or immune effector cell-associated neurotoxicity syndrome. We investigated the role of the intestinal microbiome on these outcomes in a multicenter study of patients with B cell lymphoma and leukemia. We found in a retrospective cohort (n = 228) that exposure to antibiotics, in particular piperacillin/tazobactam, meropenem and imipenem/cilastatin (P-I-M), in the 4 weeks before therapy was associated with worse survival and increased neurotoxicity. In stool samples from a prospective cohort of CAR T cell recipients (n = 48), the fecal microbiome was altered at baseline compared to healthy controls. Stool sample profiling by 16S ribosomal RNA and metagenomic shotgun sequencing revealed that clinical outcomes were associated with differences in specific bacterial taxa and metabolic pathways. Through both untargeted and hypothesis-driven analysis of 16S sequencing data, we identified species within the class Clostridia that were associated with day 100 complete response. We concluded that changes in the intestinal microbiome are associated with clinical outcomes after anti-CD19 CAR T cell therapy in patients with B cell malignancies.
© 2022. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Figures
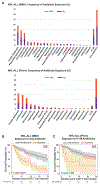





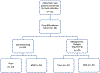
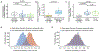

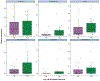



Comment in
-
Trust your gut when it comes to driving CARs.Med. 2022 May 13;3(5):281-283. doi: 10.1016/j.medj.2022.04.015. Epub 2022 May 13. Med. 2022. PMID: 35584646
References
Methods-only References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA226983/CA/NCI NIH HHS/United States
- P01 CA214278/CA/NCI NIH HHS/United States
- P01 CA023766/CA/NCI NIH HHS/United States
- R01 HL147584/HL/NHLBI NIH HHS/United States
- R01 CA219871/CA/NCI NIH HHS/United States
- R01 HL125571/HL/NHLBI NIH HHS/United States
- K99 CA212302/CA/NCI NIH HHS/United States
- K08 CA194256/CA/NCI NIH HHS/United States
- R01 CA228358/CA/NCI NIH HHS/United States
- R00 CA212302/CA/NCI NIH HHS/United States
- K08 HL143189/HL/NHLBI NIH HHS/United States
- R01 HL123340/HL/NHLBI NIH HHS/United States
- R01 CA206012/CA/NCI NIH HHS/United States
- R01 CA228308/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P01 AG052359/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

