Bridging the muscle genome to phenome across multiple biological scales
- PMID: 35288729
- PMCID: PMC9080751
- DOI: 10.1242/jeb.243630
Bridging the muscle genome to phenome across multiple biological scales
Abstract
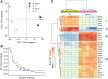
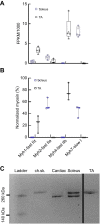
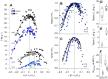
Muscle is highly hierarchically organized, with functions shaped by genetically controlled expression of protein ensembles with different isoform profiles at the sarcomere scale. However, it remains unclear how isoform profiles shape whole-muscle performance. We compared two mouse hindlimb muscles, the slow, relatively parallel-fibered soleus and the faster, more pennate-fibered tibialis anterior (TA), across scales: from gene regulation, isoform expression and translation speed, to force-length-velocity-power for intact muscles. Expression of myosin heavy-chain (MHC) isoforms directly corresponded with contraction velocity. The fast-twitch TA with fast MHC isoforms had faster unloaded velocities (actin sliding velocity, Vactin; peak fiber velocity, Vmax) than the slow-twitch soleus. For the soleus, Vactin was biased towards Vactin for purely slow MHC I, despite this muscle's even fast and slow MHC isoform composition. Our multi-scale results clearly identified a consistent and significant dampening in fiber shortening velocities for both muscles, underscoring an indirect correlation between Vactin and fiber Vmax that may be influenced by differences in fiber architecture, along with internal loading due to both passive and active effects. These influences correlate with the increased peak force and power in the slightly more pennate TA, leading to a broader length range of near-optimal force production. Conversely, a greater force-velocity curvature in the near-parallel fibered soleus highlights the fine-tuning by molecular-scale influences including myosin heavy and light chain expression along with whole-muscle characteristics. Our results demonstrate that the individual gene, protein and whole-fiber characteristics do not directly reflect overall muscle performance but that intricate fine-tuning across scales shapes specialized muscle function.
Keywords: Mouse; Myosin isoforms; Shortening velocity; Soleus; Tibialis anterior; Transcriptomics.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

