A CGA/EGFR/GATA2 positive feedback circuit confers chemoresistance in gastric cancer
- PMID: 35289315
- PMCID: PMC8920335
- DOI: 10.1172/JCI154074
A CGA/EGFR/GATA2 positive feedback circuit confers chemoresistance in gastric cancer
Abstract
De novo and acquired resistance are major impediments to the efficacy of conventional and targeted cancer therapy. In unselected gastric cancer (GC) patients with advanced disease, trials combining chemotherapy and an anti-EGFR monoclonal antibody have been largely unsuccessful. In an effort to identify biomarkers of resistance so as to better select patients for such trials, we screened the secretome of chemotherapy-treated human GC cell lines. We found that levels of CGA, the α-subunit of glycoprotein hormones, were markedly increased in the conditioned media of chemoresistant GC cells, and CGA immunoreactivity was enhanced in GC tissues that progressed on chemotherapy. CGA levels in plasma increased in GC patients who received chemotherapy, and this increase was correlated with reduced responsiveness to chemotherapy and poor survival. Mechanistically, secreted CGA was found to bind to EGFR and activate EGFR signaling, thereby conferring a survival advantage to GC cells. N-glycosylation of CGA at Asn52 and Asn78 is required for its stability, secretion, and interaction with EGFR. GATA2 was found to activate CGA transcription, whose increase, in turn, induced the expression and phosphorylation of GATA2 in an EGFR-dependent manner, forming a positive feedback circuit that was initiated by GATA2 autoregulation upon sublethal exposure to chemotherapy. Based on this circuit, combination strategies involving anti-EGFR therapies or targeting CGA with microRNAs (miR-708-3p and miR-761) restored chemotherapy sensitivity. These findings identify a clinically actionable CGA/EGFR/GATA2 circuit and highlight CGA as a predictive biomarker and therapeutic target in chemoresistant GC.
Keywords: Gastric cancer; Gastroenterology; Oncogenes; Oncology.
Conflict of interest statement
Figures
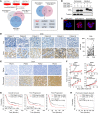
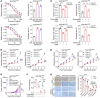

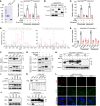

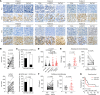

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

