Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines
- PMID: 35289637
- PMCID: PMC8995028
- DOI: 10.1126/scitranslmed.abn9243
Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines
Abstract
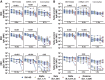
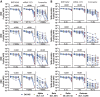
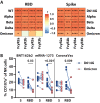
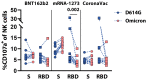
The Omicron variant of SARS-CoV-2 has been shown to evade neutralizing antibodies elicited by vaccination or infection. Despite the global spread of the Omicron variant, even among highly vaccinated populations, death rates have not increased concomitantly. These data suggest that immune mechanisms beyond antibody-mediated virus neutralization may protect against severe disease. In addition to neutralizing pathogens, antibodies contribute to control and clearance of infections through Fc effector mechanisms. Here, we probed the ability of vaccine-induced antibodies to drive Fc effector activity against the Omicron variant using samples from individuals receiving one of three SARS-CoV-2 vaccines. Despite a substantial loss of IgM, IgA, and IgG binding to the Omicron variant receptor binding domain (RBD) in samples from individuals receiving BNT162b2, mRNA-1273, and CoronaVac vaccines, stable binding was maintained against the full-length Omicron Spike protein. Compromised RBD binding IgG was accompanied by a loss of RBD-specific antibody Fcγ receptor (FcγR) binding in samples from individuals who received the CoronaVac vaccine, but RBD-specific FcγR2a and FcγR3a binding was preserved in recipients of mRNA vaccines. Conversely, Spike protein-specific antibodies exhibited persistent but reduced binding to FcγRs across all three vaccines, although higher binding was observed in samples from recipients of mRNA vaccines. This was associated with preservation of FcγR2a and FcγR3a binding antibodies and maintenance of Spike protein-specific antibody-dependent natural killer cell activation. Thus, despite the loss of Omicron neutralization, vaccine-induced Spike protein-specific antibodies continue to drive Fc effector functions, suggesting a capacity for extraneutralizing antibodies to contribute to disease control.
Figures
Update of
-
Preserved Omicron Spike specific antibody binding and Fc-recognition across COVID-19 vaccine platforms.medRxiv [Preprint]. 2021 Dec 27:2021.12.24.21268378. doi: 10.1101/2021.12.24.21268378. medRxiv. 2021. Update in: Sci Transl Med. 2022 Apr 27;14(642):eabn9243. doi: 10.1126/scitranslmed.abn9243. PMID: 34981072 Free PMC article. Updated. Preprint.
References
-
- Khoury D. S., Cromer D., Reynaldi A., Schlub T. E., Wheatley A. K., Juno J. A., Subbarao K., Kent S. J., Triccas J. A., Davenport M. P., Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021). 10.1038/s41591-021-01377-8 - DOI - PubMed
-
- Peacock T. P., Brown J. C., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W. S., The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv, 2021.2012.2031.474653 (2022). 10.1101/2021.12.31.474653 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- U19 AI135995/AI/NIAID NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- R01 AI042790/AI/NIAID NIH HHS/United States
- R01 AI146785/AI/NIAID NIH HHS/United States
- R01 AI147884/AI/NIAID NIH HHS/United States
- UM1 AI148373/AI/NIAID NIH HHS/United States
- HHSN272201500002C/AI/NIAID NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- 75N93019C00052/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- UM1 AI148576/AI/NIAID NIH HHS/United States
- U01 CA260476/CA/NCI NIH HHS/United States
- R37 AI080289/AI/NIAID NIH HHS/United States
- U01 AI149644/AI/NIAID NIH HHS/United States
- UM1 AI148684/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

