Single-cell transcriptomics reveals distinct effector profiles of infiltrating T cells in lupus skin and kidney
- PMID: 35290245
- PMCID: PMC9089784
- DOI: 10.1172/jci.insight.156341
Single-cell transcriptomics reveals distinct effector profiles of infiltrating T cells in lupus skin and kidney
Abstract
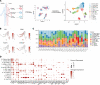
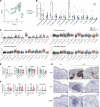
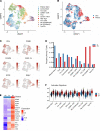
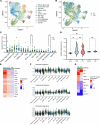
Cutaneous lupus is commonly present in patients with systemic lupus erythematosus (SLE). T cells have been strongly suspected to contribute to the pathology of cutaneous lupus; however, our understanding of the relevant T cell phenotypes and functions remains incomplete. Here, we present a detailed single-cell RNA-Seq profile of T and NK cell populations present within lesional and nonlesional skin biopsies of patients with cutaneous lupus. T cells across clusters from lesional and nonlesional skin biopsies expressed elevated levels of IFN-simulated genes (ISGs). Compared with T cells from control skin, however, T cells from cutaneous lupus lesions did not show elevated expression profiles of activation, cytotoxicity, or exhaustion. Integrated analyses indicated that skin lymphocytes appeared less activated and lacked the expanded cytotoxic populations prominent in lupus nephritis kidney T/NK cells. Comparison of skin T cells from lupus and systemic sclerosis skin biopsies further revealed an elevated ISG signature specific to cells from lupus biopsies. Overall, these data represent the first detailed transcriptomic analysis to our knowledge of the T and NK cells in cutaneous lupus at the single-cell level and have enabled a cross-tissue comparison that highlights stark differences in composition and activation of T/NK cells in distinct tissues in lupus.
Keywords: Adaptive immunity; Autoimmunity; Skin; T cells.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

