Gene selection using pyramid gravitational search algorithm
- PMID: 35290401
- PMCID: PMC8923457
- DOI: 10.1371/journal.pone.0265351
Gene selection using pyramid gravitational search algorithm
Abstract
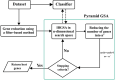
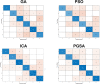
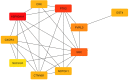
Genetics play a prominent role in the development and progression of malignant neoplasms. Identification of the relevant genes is a high-dimensional data processing problem. Pyramid gravitational search algorithm (PGSA), a hybrid method in which the number of genes is cyclically reduced is proposed to conquer the curse of dimensionality. PGSA consists of two elements, a filter and a wrapper method (inspired by the gravitational search algorithm) which iterates through cycles. The genes selected in each cycle are passed on to the subsequent cycles to further reduce the dimension. PGSA tries to maximize the classification accuracy using the most informative genes while reducing the number of genes. Results are reported on a multi-class microarray gene expression dataset for breast cancer. Several feature selection algorithms have been implemented to have a fair comparison. The PGSA ranked first in terms of accuracy (84.5%) with 73 genes. To check if the selected genes are meaningful in terms of patient's survival and response to therapy, protein-protein interaction network analysis has been applied on the genes. An interesting pattern was emerged when examining the genetic network. HSP90AA1, PTK2 and SRC genes were amongst the top-rated bottleneck genes, and DNA damage, cell adhesion and migration pathways are highly enriched in the network.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Development of a two-stage gene selection method that incorporates a novel hybrid approach using the cuckoo optimization algorithm and harmony search for cancer classification.J Biomed Inform. 2017 Mar;67:11-20. doi: 10.1016/j.jbi.2017.01.016. Epub 2017 Feb 3. J Biomed Inform. 2017. PMID: 28163197
-
Regularization network-based gene selection for microarray data analysis.Int J Neural Syst. 2006 Oct;16(5):341-52. doi: 10.1142/S0129065706000743. Int J Neural Syst. 2006. PMID: 17117495
-
Improving accuracy for cancer classification with a new algorithm for genes selection.BMC Bioinformatics. 2012 Nov 13;13:298. doi: 10.1186/1471-2105-13-298. BMC Bioinformatics. 2012. PMID: 23148517 Free PMC article.
-
A TRIZ-inspired bat algorithm for gene selection in cancer classification.Genomics. 2020 Jan;112(1):114-126. doi: 10.1016/j.ygeno.2019.09.015. Epub 2019 Oct 30. Genomics. 2020. PMID: 31676302
-
Filter versus wrapper gene selection approaches in DNA microarray domains.Artif Intell Med. 2004 Jun;31(2):91-103. doi: 10.1016/j.artmed.2004.01.007. Artif Intell Med. 2004. PMID: 15219288 Review.
Cited by
-
Feature selection for high dimensional microarray gene expression data via weighted signal to noise ratio.PLoS One. 2023 Apr 25;18(4):e0284619. doi: 10.1371/journal.pone.0284619. eCollection 2023. PLoS One. 2023. PMID: 37098036 Free PMC article.
-
Optimized models and deep learning methods for drug response prediction in cancer treatments: a review.PeerJ Comput Sci. 2024 Mar 25;10:e1903. doi: 10.7717/peerj-cs.1903. eCollection 2024. PeerJ Comput Sci. 2024. PMID: 38660174 Free PMC article.
-
A Hybrid Machine Learning Approach to Screen Optimal Predictors for the Classification of Primary Breast Tumors from Gene Expression Microarray Data.Diagnostics (Basel). 2023 Feb 13;13(4):708. doi: 10.3390/diagnostics13040708. Diagnostics (Basel). 2023. PMID: 36832196 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al.. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. 2021;71(3):209–49. - PubMed
-
- Masuda H, Masuda N, Kodama Y, Ogawa M, Karita M, Yamamura J, et al.. Predictive factors for the effectiveness of neoadjuvant chemotherapy and prognosis in triple-negative breast cancer patients. Cancer Chemotherapy and Pharmacology 2010 67:4. 2010;67(4):911–7. doi: 10.1007/s00280-010-1371-4 - DOI - PubMed
-
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al.. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor–Positive Breast Cancer. Clinical Cancer Research. 2010;16(21):5222–32. doi: 10.1158/1078-0432.CCR-10-1282 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

