Intrinsically disordered signaling proteins: Essential hub players in the control of stress responses in Saccharomyces cerevisiae
- PMID: 35290420
- PMCID: PMC8923507
- DOI: 10.1371/journal.pone.0265422
Intrinsically disordered signaling proteins: Essential hub players in the control of stress responses in Saccharomyces cerevisiae
Abstract
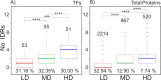
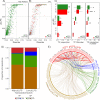
Cells have developed diverse mechanisms to monitor changes in their surroundings. This allows them to establish effective responses to cope with adverse environments. Some of these mechanisms have been well characterized in the budding yeast Saccharomyces cerevisiae, an excellent experimental model to explore and elucidate some of the strategies selected in eukaryotic organisms to adjust their growth and development in stressful conditions. The relevance of structural disorder in proteins and the impact on their functions has been uncovered for proteins participating in different processes. This is the case of some transcription factors (TFs) and other signaling hub proteins, where intrinsically disordered regions (IDRs) play a critical role in their function. In this work, we present a comprehensive bioinformatic analysis to evaluate the significance of structural disorder in those TFs (170) recognized in S. cerevisiae. Our findings show that 85.2% of these TFs contain at least one IDR, whereas ~30% exhibit a higher disorder level and thus were considered as intrinsically disordered proteins (IDPs). We also found that TFs contain a higher number of IDRs compared to the rest of the yeast proteins, and that intrinsically disordered TFs (IDTFs) have a higher number of protein-protein interactions than those with low structural disorder. The analysis of different stress response pathways showed a high content of structural disorder not only in TFs but also in other signaling proteins. The propensity of yeast proteome to undergo a liquid-liquid phase separation (LLPS) was also analyzed, showing that a significant proportion of IDTFs may undergo this phenomenon. Our analysis is a starting point for future research on the importance of structural disorder in yeast stress responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
The Balancing Act of Intrinsically Disordered Proteins: Enabling Functional Diversity while Minimizing Promiscuity.J Mol Biol. 2019 Apr 5;431(8):1650-1670. doi: 10.1016/j.jmb.2019.03.008. Epub 2019 Mar 13. J Mol Biol. 2019. PMID: 30878482 Free PMC article.
-
Intrinsically Disordered Regions Direct Transcription Factor In Vivo Binding Specificity.Mol Cell. 2020 Aug 6;79(3):459-471.e4. doi: 10.1016/j.molcel.2020.05.032. Epub 2020 Jun 16. Mol Cell. 2020. PMID: 32553192
-
The molecular grammar of protein disorder guiding genome-binding locations.Nucleic Acids Res. 2023 Jun 9;51(10):4831-4844. doi: 10.1093/nar/gkad184. Nucleic Acids Res. 2023. PMID: 36938874 Free PMC article.
-
Lighting up Nobel Prize-winning studies with protein intrinsic disorder.Cell Mol Life Sci. 2022 Jul 26;79(8):449. doi: 10.1007/s00018-022-04468-y. Cell Mol Life Sci. 2022. PMID: 35882686 Free PMC article. Review.
-
Functional unfoldomics: Roles of intrinsic disorder in protein (multi)functionality.Adv Protein Chem Struct Biol. 2024;138:179-210. doi: 10.1016/bs.apcsb.2023.11.001. Epub 2023 Nov 22. Adv Protein Chem Struct Biol. 2024. PMID: 38220424 Review.
Cited by
-
Enrichment patterns of intrinsic disorder in proteins.Biophys Rev. 2022 Nov 19;14(6):1487-1493. doi: 10.1007/s12551-022-01016-7. eCollection 2022 Dec. Biophys Rev. 2022. PMID: 36659984 Free PMC article. Review.
-
Targeting stressor-induced dysfunctions in protein-protein interaction networks via epichaperomes.Trends Pharmacol Sci. 2023 Jan;44(1):20-33. doi: 10.1016/j.tips.2022.10.006. Epub 2022 Nov 20. Trends Pharmacol Sci. 2023. PMID: 36414432 Free PMC article. Review.
-
The GET pathway is a major bottleneck for maintaining proteostasis in Saccharomyces cerevisiae.Sci Rep. 2023 Jun 7;13(1):9285. doi: 10.1038/s41598-023-35666-8. Sci Rep. 2023. PMID: 37286562 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

