This is a preprint.
Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting
- PMID: 35291297
- PMCID: PMC8923105
- DOI: 10.1101/2022.03.08.483429
Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting
Update in
-
Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting.Antiviral Res. 2022 Dec;208:105452. doi: 10.1016/j.antiviral.2022.105452. Epub 2022 Oct 29. Antiviral Res. 2022. PMID: 36341734 Free PMC article.
Abstract
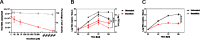
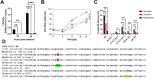
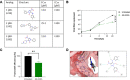
SARS-CoV-2 is currently causing an unprecedented pandemic. While vaccines are massively deployed, we still lack effective large-scale antiviral therapies. In the quest for antivirals targeting conserved structures, we focused on molecules able to bind viral RNA secondary structures. Aminoglycosides are a class of antibiotics known to interact with the ribosomal RNA of both prokaryotes and eukaryotes and have previously been shown to exert antiviral activities by interacting with viral RNA. Here we show that the aminoglycoside geneticin is endowed with antiviral activity against all tested variants of SARS-CoV-2, in different cell lines and in a respiratory tissue model at non-toxic concentrations. The mechanism of action is an early inhibition of RNA replication and protein expression related to a decrease in the efficiency of the -1 programmed ribosomal frameshift (PRF) signal of SARS-CoV-2. Using in silico modelling, we have identified a potential binding site of geneticin in the pseudoknot of frameshift RNA motif. Moreover, we have selected, through virtual screening, additional RNA binding compounds, interacting with the same site with increased potency.
Figures


Similar articles
-
Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting.Antiviral Res. 2022 Dec;208:105452. doi: 10.1016/j.antiviral.2022.105452. Epub 2022 Oct 29. Antiviral Res. 2022. PMID: 36341734 Free PMC article.
-
Targeting Ribosomal Frameshifting as an Antiviral Strategy: From HIV-1 to SARS-CoV-2.Acc Chem Res. 2021 Sep 7;54(17):3349-3361. doi: 10.1021/acs.accounts.1c00316. Epub 2021 Aug 17. Acc Chem Res. 2021. PMID: 34403258 Review.
-
Anti-Frameshifting Ligand Active against SARS Coronavirus-2 Is Resistant to Natural Mutations of the Frameshift-Stimulatory Pseudoknot.J Mol Biol. 2020 Oct 2;432(21):5843-5847. doi: 10.1016/j.jmb.2020.09.006. Epub 2020 Sep 11. J Mol Biol. 2020. PMID: 32920049 Free PMC article.
-
Restriction of SARS-CoV-2 Replication by Targeting Programmed -1 Ribosomal Frameshifting In Vitro.bioRxiv [Preprint]. 2020 Oct 21:2020.10.21.349225. doi: 10.1101/2020.10.21.349225. bioRxiv. 2020. Update in: Proc Natl Acad Sci U S A. 2021 Jun 29;118(26):e2023051118. doi: 10.1073/pnas.2023051118. PMID: 33106809 Free PMC article. Updated. Preprint.
-
Small Molecules Targeting Viral RNA.Int J Mol Sci. 2023 Aug 31;24(17):13500. doi: 10.3390/ijms241713500. Int J Mol Sci. 2023. PMID: 37686306 Free PMC article. Review.
References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindah E. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. 10.1016/j.softx.2015.06.001. - DOI
-
- Ahn D.G., Lee W., Choi J.K., Kim S.J., Plant E.P., Almazán F., Taylor D.R., Enjuanes L., and Oh J.W. (2011). Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antiviral Research 91, 1–10. 10.1016/j.antiviral.2011.04.009. - DOI - PMC - PubMed
-
- Birk1 Alexander V, Dubovi Edward J, Zhang Xianchao, and Szeto Hazel H(2008). Antiviral activity of geneticin against bovine viral diarrhoea virus. Antiviral Chemistry & Chemotherapy 19, 33–40. - PubMed
-
- Ariza-Mateos A., Díaz-Toledano R., Block T.M., Prieto-Vega S., Birk A., and Gómez J. (2016). Geneticin stabilizes the open conformation of the 5′ region of hepatitis C virus RNA and inhibits viral replication. Antimicrobial Agents and Chemotherapy 60, 925–935. 10.1128/AAC.02511-15. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
