This is a preprint.
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion
- PMID: 35291301
- PMCID: PMC8923104
- DOI: 10.1101/2022.03.04.483074
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion
Update in
-
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion.PLoS Pathog. 2025 Jun 20;21(6):e1013295. doi: 10.1371/journal.ppat.1013295. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40540531 Free PMC article.
Abstract
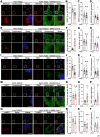
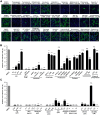
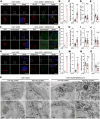
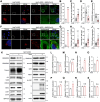
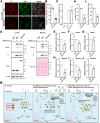
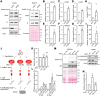
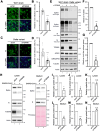
The COVID-19 pandemic is caused by SARS-CoV-2, an enveloped RNA virus. Despite extensive investigation, the molecular mechanisms for its assembly and secretion remain largely elusive. Here, we show that SARS-CoV-2 infection induces global alterations of the host endomembrane system, including dramatic Golgi fragmentation. SARS-CoV-2 virions are enriched in the fragmented Golgi. Disrupting Golgi function with small molecules strongly inhibits viral infection. Significantly, SARS-CoV-2 infection down-regulates GRASP55 but up-regulates TGN46 protein levels. Surprisingly, GRASP55 expression reduces both viral secretion and spike number on each virion, while GRASP55 depletion displays opposite effects. In contrast, TGN46 depletion only inhibits viral secretion without affecting spike incorporation into virions. TGN46 depletion and GRASP55 expression additively inhibit viral secretion, indicating that they act at different stages. Taken together, we show that SARS-CoV-2 alters Golgi structure and function to control viral assembly and secretion, highlighting the Golgi as a potential therapeutic target for blocking SARS-CoV-2 infection.
Keywords: GRASP55; Golgi fragmentation; SARS-CoV-2; TGN46; trafficking; viral assembly.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
