IL-4 polarized human macrophage exosomes control cardiometabolic inflammation and diabetes in obesity
- PMID: 35292359
- PMCID: PMC9171286
- DOI: 10.1016/j.ymthe.2022.03.008
IL-4 polarized human macrophage exosomes control cardiometabolic inflammation and diabetes in obesity
Abstract
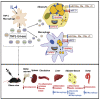
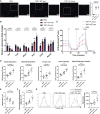
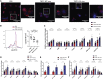
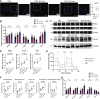
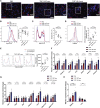
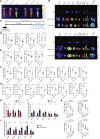
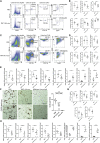
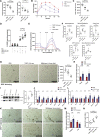
Cardiometabolic disease is an increasing cause of morbidity and death in society. While M1-like macrophages contribute to metabolic inflammation and insulin resistance, those polarized to an M2-like phenotype exert protective properties. Building on our observations reporting M2-like macrophage exosomes in atherosclerosis control, we tested whether they could serve to control inflammation in the liver and adipose tissue of obese mice. In thinking of clinical translation, we studied human THP-1 macrophages exposed to interleukin (IL)-4 as a source of exosomes (THP1-IL4-exo). Our findings show that THP1-IL4-exo polarized primary macrophages to an anti-inflammatory phenotype and reprogramed their energy metabolism by increasing levels of microRNA-21/99a/146b/378a (miR-21/99a/146b/378a) while reducing miR-33. This increased lipophagy, mitochondrial activity, and oxidative phosphorylation (OXPHOS). THP1-IL4-exo exerted a similar regulation of these miRs in cultured 3T3-L1 adipocytes. This enhanced insulin-dependent glucose uptake through increased peroxisome proliferator activated receptor gamma (PPARγ)-driven expression of GLUT4. It also increased levels of UCP1 and OXPHOS activity, which promoted lipophagy, mitochondrial activity, and beiging of 3T3-L1 adipocytes. Intraperitoneal infusions of THP1-IL4-exo into obese wild-type and Ldlr-/- mice fed a Western high-fat diet reduced hematopoiesis and myelopoiesis, and favorably reprogramed inflammatory signaling and metabolism in circulating Ly6Chi monocytes. This also reduced leukocyte numbers and inflammatory activity in the circulation, aorta, adipose tissue, and the liver. Such treatments reduced hepatic steatosis and increased the beiging of white adipose tissue as revealed by increased UCP1 expression and OXPHOS activity that normalized blood insulin levels and improved glucose tolerance. Our findings support THP1-IL4-exo as a therapeutic approach to control cardiometabolic disease and diabetes in obesity.
Keywords: PPARγ; adipocyte; beiging; cardiometabolic inflammation; exosomes; macrophage; microRNA-33; mitochondrial respiration; obesity; type II diabetes.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests T.A.P., M.N., N.K.V., L.B., and R.L.R. have filed an invention disclosure related to some aspects of this work with the University of California, San Francisco, and the US Department of Veterans Affairs.
Figures

References
-
- Lumeng C.N., Deyoung S.M., Bodzin J.L., Saltiel A.R. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. - PubMed

