Microtubule rescue at midzone edges promotes overlap stability and prevents spindle collapse during anaphase B
- PMID: 35293864
- PMCID: PMC9018073
- DOI: 10.7554/eLife.72630
Microtubule rescue at midzone edges promotes overlap stability and prevents spindle collapse during anaphase B
Abstract
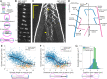
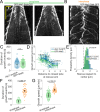
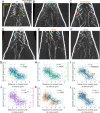
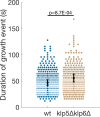
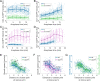
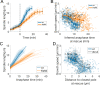
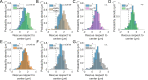
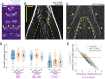
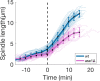
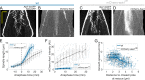
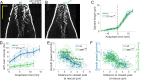
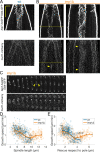
During anaphase B, molecular motors slide interpolar microtubules to elongate the mitotic spindle, contributing to the separation of chromosomes. However, sliding of antiparallel microtubules reduces their overlap, which may lead to spindle breakage, unless microtubules grow to compensate sliding. How sliding and growth are coordinated is still poorly understood. In this study, we have used the fission yeast S. pombe to measure microtubule dynamics during anaphase B. We report that the coordination of microtubule growth and sliding relies on promoting rescues at the midzone edges. This makes microtubules stable from pole to midzone, while their distal parts including the plus ends alternate between assembly and disassembly. Consequently, the midzone keeps a constant length throughout anaphase, enabling sustained sliding without the need for a precise regulation of microtubule growth speed. Additionally, we found that in S. pombe, which undergoes closed mitosis, microtubule growth speed decreases when the nuclear membrane wraps around the spindle midzone.
Keywords: S. pombe; anaphase; cell biology; live cell imaging; microtubule dynamics; microtubule sliding; modelling; spindle.
© 2022, Lera-Ramirez et al.
Conflict of interest statement
ML, FN, PT No competing interests declared
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

