MtGSTF7, a TT19-like GST gene, is essential for accumulation of anthocyanins, but not proanthocyanins in Medicago truncatula
- PMID: 35294003
- PMCID: PMC9232208
- DOI: 10.1093/jxb/erac112
MtGSTF7, a TT19-like GST gene, is essential for accumulation of anthocyanins, but not proanthocyanins in Medicago truncatula
Abstract
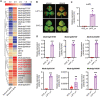
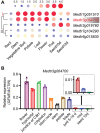
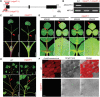
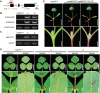
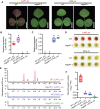
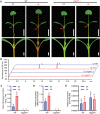
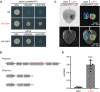
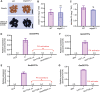
Anthocyanins and proanthocyanins (PAs) are two end products of the flavonoid biosynthesis pathway. They are believed to be synthesized in the endoplasmic reticulum and then sequestered into the vacuole. In Arabidopsis thaliana, TRANSPARENT TESTA 19 (TT19) is necessary for both anthocyanin and PA accumulation. Here, we found that MtGSTF7, a homolog of AtTT19, is essential for anthocyanin accumulation but not required for PA accumulation in Medicago truncatula. MtGSTF7 was induced by the anthocyanin regulator LEGUME ANTHOCYANIN PRODUCTION 1 (LAP1), and its tissue expression pattern correlated with anthocyanin deposition in M. truncatula. Tnt1-insertional mutants of MtGSTF7 lost anthocyanin accumulation in vegetative organs, and introducing a genomic fragment of MtGSTF7 could complement the mutant phenotypes. Additionally, the accumulation of anthocyanins induced by LAP1 was significantly reduced in mtgstf7 mutants. Yeast-one-hybridization and dual-luciferase reporter assays revealed that LAP1 could bind to the MtGSTF7 promoter to activate its expression. Ectopic expression of MtGSTF7 in tt19 mutants could rescue their anthocyanin deficiency, but not their PA defect. Furthermore, PA accumulation was not affected in the mtgstf7 mutants. Taken together, our results show that the mechanism of anthocyanin and PA accumulation in M. truncatula is different from that in A. thaliana, and provide a new target gene for engineering anthocyanins in plants.
Keywords: Medicago truncatula; Anthocyanin; LAP1; MtGSTF7; glutathione-S-transferase; proanthocyanidin.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Similar articles
-
Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8.New Phytol. 2016 May;210(3):905-21. doi: 10.1111/nph.13816. Epub 2016 Jan 4. New Phytol. 2016. PMID: 26725247
-
The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago.Plant J. 2009 Jul;59(1):136-49. doi: 10.1111/j.1365-313X.2009.03885.x. Epub 2009 Apr 2. Plant J. 2009. PMID: 19368693
-
A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development.Plant Physiol. 2009 Nov;151(3):1114-29. doi: 10.1104/pp.109.144022. Epub 2009 Aug 26. Plant Physiol. 2009. PMID: 19710231 Free PMC article.
-
Model Legumes: Functional Genomics Tools in Medicago truncatula.Methods Mol Biol. 2018;1822:11-37. doi: 10.1007/978-1-4939-8633-0_2. Methods Mol Biol. 2018. PMID: 30043294 Review.
-
Anthocyanin stability and degradation in plants.Plant Signal Behav. 2021 Dec 2;16(12):1987767. doi: 10.1080/15592324.2021.1987767. Epub 2021 Oct 22. Plant Signal Behav. 2021. PMID: 34686106 Free PMC article. Review.
Cited by
-
The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses.Plants (Basel). 2023 Jul 5;12(13):2558. doi: 10.3390/plants12132558. Plants (Basel). 2023. PMID: 37447119 Free PMC article. Review.
-
Transcription factor NnMYB5 controls petal color by regulating GLUTATHIONE S-TRANSFERASE2 in Nelumbo nucifera.Plant Physiol. 2023 Sep 22;193(2):1213-1226. doi: 10.1093/plphys/kiad363. Plant Physiol. 2023. PMID: 37348874 Free PMC article.
-
The flexibility of proanthocyanidin biosynthesis in plants.Plant Physiol. 2022 Aug 29;190(1):202-205. doi: 10.1093/plphys/kiac274. Plant Physiol. 2022. PMID: 35695780 Free PMC article.
-
Downregulation of a Phi class glutathione S-transferase gene in transgenic torenia yielded pale flower color.Plant Biotechnol (Tokyo). 2024 Jun 25;41(2):147-151. doi: 10.5511/plantbiotechnology.24.0409a. Plant Biotechnol (Tokyo). 2024. PMID: 39463771 Free PMC article.
-
Overlooked and misunderstood: can glutathione conjugates be clues to understanding plant glutathione transferases?Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230365. doi: 10.1098/rstb.2023.0365. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343017 Free PMC article. Review.
References
-
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM.. 2011. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65, 771–784. - PubMed
-
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. . 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. - PubMed
-
- Braidot E, Petrussa E, Bertolini A, Peresson C, Ermacora P, Loi N, Terdoslavich M, Passamonti S, Macrì F, Vianello A.. 2008a. Evidence for a putative flavonoid translocator similar to mammalian bilitranslocase in grape berries (Vitis vinifera L.) during ripening. Planta 228, 203–213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- XDA26030301/Strategic Priority Research Program of the Chinese Academy of Sciences
- U1702234/National Natural Science Foundation of China
- 2015HA032/High-end Scientific and Technological Talents in Yunnan Province
- 202101AW070004/Yunnan Fundamental Research Projects
- 2021395/Youth Innovation Promotion Association CAS
LinkOut - more resources
Full Text Sources
Research Materials

