Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages
- PMID: 35294555
- PMCID: PMC8926390
- DOI: 10.1093/molbev/msac045
Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages
Abstract
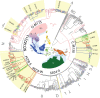
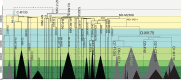
Island Southeast Asia (ISEA) and Oceania host one of the world's richest assemblages of human phenotypic, linguistic, and cultural diversity. Despite this, the region's male genetic lineages are globally among the last to remain unresolved. We compiled ∼9.7 Mb of Y chromosome (chrY) sequence from a diverse sample of over 380 men from this region, including 152 first reported here. The granularity of this data set allows us to fully resolve and date the regional chrY phylogeny. This new high-resolution tree confirms two main population bursts: multiple rapid diversifications following the region's initial settlement ∼50 kya, and extensive expansions <6 kya. Notably, ∼40-25 kya the deep rooting local lineages of C-M130, M-P256, and S-B254 show almost no further branching events in ISEA, New Guinea, and Australia, matching a similar pause in diversification seen in maternal mitochondrial DNA lineages. The main local lineages start diversifying ∼25 kya, at the time of the last glacial maximum. This improved chrY topology highlights localized events with important historical implications, including pre-Holocene contact between Mainland and ISEA, potential interactions between Australia and the Papuan world, and a sustained period of diversification following the flooding of the ancient Sunda and Sahul continents as the insular landscape observed today formed. The high-resolution phylogeny of the chrY presented here thus enables a detailed exploration of past isolation, interaction, and change in one of the world's least understood regions.
Keywords: Island Southeast Asia; Y chromosome; human population genetics; migration; phylogeography.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Beckett J. 1987. Torres Strait Islanders: custom and colonialism. Cambridge: Cambridge University Press.
-
- Bellwood P. 2004. First farmers: the origins of agricultural societies. New York: Wiley.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

