Distinct Expansion of Group II Introns During Evolution of Prokaryotes and Possible Factors Involved in Its Regulation
- PMID: 35295308
- PMCID: PMC8919778
- DOI: 10.3389/fmicb.2022.849080
Distinct Expansion of Group II Introns During Evolution of Prokaryotes and Possible Factors Involved in Its Regulation
Abstract
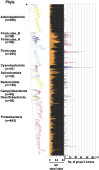
Group II introns (G2Is) are ribozymes that have retroelement characteristics in prokaryotes. Although G2Is are suggested to have been an important evolutionary factor in the prokaryote-to-eukaryote transition, comprehensive analyses of these introns among the tens of thousands of prokaryotic genomes currently available are still limited. Here, we developed a bioinformatic pipeline that systematically collects G2Is and applied it to prokaryotic genomes. We found that in bacteria, 25% (447 of 1,790) of the total representative genomes had an average of 5.3 G2Is, and in archaea, 9% (28 of 296) of the total representative genomes had an average of 3.0 G2Is. The greatest number of G2Is per genome was 101 in Arthrospira platensis (phylum Cyanobacteriota). A comprehensive sequence analysis of the intron-encoded protein (IEP) in each G2I sequence was conducted and resulted in the addition of three new IEP classes (U1-U3) to the previous classification. This analysis suggested that about 30% of all IEPs are non-canonical IEPs. The number of G2Is per genome was defined almost at the phylum level, and at least in the following two phyla, Firmicutes, and Cyanobacteriota, the type of IEP was largely associated as a factor in the G2I increase, i.e., there was an explosive increase in G2Is with bacterial C-type IEPs, mainly in the phylum Firmicutes, and in G2Is with CL-type IEPs, mainly in the phylum Cyanobacteriota. We also systematically analyzed the relationship between genomic signatures and the mechanism of these increases in G2Is. This is the first study to systematically characterize G2Is in the prokaryotic phylogenies.
Keywords: bioinformatics; genomic signatures; group II intron; intron-encoded protein; prokaryotic genomes.
Copyright © 2022 Miura, Nagata, Tamaki, Tomita and Kanai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
A pipeline of programs for collecting and analyzing group II intron retroelement sequences from GenBank.Mob DNA. 2013 Dec 20;4(1):28. doi: 10.1186/1759-8753-4-28. Mob DNA. 2013. PMID: 24359548 Free PMC article.
-
Insights into the strategies used by related group II introns to adapt successfully for the colonisation of a bacterial genome.RNA Biol. 2014;11(8):1061-71. doi: 10.4161/rna.32092. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482895 Free PMC article. Review.
-
An intronic open reading frame was released from one of group II introns in the mitochondrial genome of the haptophyte Chrysochromulina sp. NIES-1333.Mob Genet Elements. 2014 May 27;4:e29384. doi: 10.4161/mge.29384. eCollection 2014. Mob Genet Elements. 2014. PMID: 25054084 Free PMC article.
-
Recent horizontal transfer, functional adaptation and dissemination of a bacterial group II intron.BMC Evol Biol. 2016 Oct 20;16(1):223. doi: 10.1186/s12862-016-0789-7. BMC Evol Biol. 2016. PMID: 27765015 Free PMC article.
-
Group II introns in the bacterial world.Mol Microbiol. 2000 Dec;38(5):917-26. doi: 10.1046/j.1365-2958.2000.02197.x. Mol Microbiol. 2000. PMID: 11123668 Review.
Cited by
-
Plant organellar RNA maturation.Plant Cell. 2023 May 29;35(6):1727-1751. doi: 10.1093/plcell/koad049. Plant Cell. 2023. PMID: 36807982 Free PMC article. Review.
-
Presence of group II introns in phage genomes.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf761. doi: 10.1093/nar/gkaf761. Nucleic Acids Res. 2025. PMID: 40808305 Free PMC article.
-
Possible Acquisition and Molecular Evolution of vpu Genes Inferred from Comprehensive Sequence Analysis of Human and Simian Immunodeficiency Viruses.J Mol Evol. 2025 Aug;93(4):478-493. doi: 10.1007/s00239-025-10256-6. Epub 2025 Jun 21. J Mol Evol. 2025. PMID: 40544231 Free PMC article.
-
Prevalence of Group II Introns in Phage Genomes.bioRxiv [Preprint]. 2025 May 23:2025.05.22.655115. doi: 10.1101/2025.05.22.655115. bioRxiv. 2025. Update in: Nucleic Acids Res. 2025 Aug 11;53(15):gkaf761. doi: 10.1093/nar/gkaf761. PMID: 40475605 Free PMC article. Updated. Preprint.
-
Group II Intron-Encoded Proteins (IEPs/Maturases) as Key Regulators of Nad1 Expression and Complex I Biogenesis in Land Plant Mitochondria.Genes (Basel). 2022 Jun 24;13(7):1137. doi: 10.3390/genes13071137. Genes (Basel). 2022. PMID: 35885919 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

