EST-SSR Primer Development and Genetic Structure Analysis of Psathyrostachys juncea Nevski
- PMID: 35295628
- PMCID: PMC8919075
- DOI: 10.3389/fpls.2022.837787
EST-SSR Primer Development and Genetic Structure Analysis of Psathyrostachys juncea Nevski
Abstract
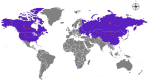
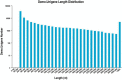
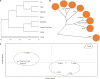
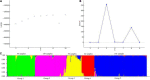
Psathyrostachys juncea is a perennial forage grass which plays an important role in soil and water conservation and ecological maintenance in cold and dry areas of temperate regions. In P. juncea, a variety of biotic and abiotic stress related genes have been used in crop improvement, indicating its agronomic, economic, forage, and breeding value. To date, there have been few studies on the genetic structure of P. juncea. Here, the genetic diversity and population structure of P. juncea were analyzed by EST-SSR molecular markers to evaluate the genetic differentiation related to tillering traits in P. juncea germplasm resources. The results showed that 400 simple sequence repeat (SSR) loci were detected in 2,020 differentially expressed tillering related genes. A total of 344 scored bands were amplified using 103 primer pairs, out of which 308 (89.53%) were polymorphic. The Nei's gene diversity of 480 individuals was between 0.092 and 0.449, and the genetic similarity coefficient was between 0.5008 and 0.9111, with an average of 0.6618. Analysis of molecular variance analysis showed that 93% of the variance was due to differences within the population, and the remaining 7% was due to differences among populations. Psathyrostachys juncea materials were clustered into five groups based on population genetic structure, principal coordinate analysis and unweighted pair-group method with arithmetic means (UPGMA) analysis. The results were similar between clustering methods, but a few individual plants were distributed differently by the three models. The clustering results, gene diversity and genetic similarity coefficients showed that the overall genetic relationship of P. juncea individuals was relatively close. A Mantel test, UPGMA and structural analysis also showed a significant correlation between genetic relationship and geographical distribution. These results provide references for future breeding programs, genetic improvement and core germplasm collection of P. juncea.
Keywords: EST-SSR; Psathyrostachys juncea Nevski; forage grass; genetic diversity; population structure.
Copyright © 2022 Li, Yun, Gao, Wang, Ren and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Athipat N., Worawit S., Katanyutita D., Apinya L., Kate-Ngam S., Jantasuriyarat C., et al. (2019). Substantial enhancement of high polymorphic SSR marker development using in silico method from 18 available rice blast fungus genome sequences and its application in genetic diversity assessment. Biologia 74 1181–1189. 10.2478/s11756-019-00264-5 - DOI
-
- Bolaric S., Barth S., Melchinger A. E., Posselt U. K. (2005). Genetic diversity in European perennial ryegrass cultivars investigated with RAPD markers. Plant Breed. 124 161–166. 10.1111/j.1439-0523.2004.01032 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

