Development and Validation of a Novel and Fast Detection Method for Cannabis sativa: A 19-Plex Short Tandem Repeat Typing System
- PMID: 35295633
- PMCID: PMC8918947
- DOI: 10.3389/fpls.2022.837945
Development and Validation of a Novel and Fast Detection Method for Cannabis sativa: A 19-Plex Short Tandem Repeat Typing System
Abstract
In recent years, influenced by the legalization of Cannabis sativa in some countries and regions, the number of people who smoke or abuse C. sativa has continuously grown, cases of transnational C. sativa trafficking have also been increasing. Therefore, fast and accurate identification and source tracking of C. sativa have become urgent social needs. In this study, we developed a new 19-plex short tandem repeats (STRs) typing system for C. sativa, which includes 15 autosomal STRs (D02-CANN1, C11-CANN1, 4910, B01-CANN1, E07-CANN1, 9269, B05-CANN1, H06-CANN2, 5159, nH09, CS1, ANUCS 305, 3735, and ANUCS 302 and 9043), two X-chromosome STRs (ANUCS 501 and 1528), one sex-determining marker (DM016, on Y-chromosome), and a quality control marker (DM029, on autosome). The whole polymerase chain reaction (PCR) process could finish within 1 h, making the system suitable for fast detection. The PCR products were detected and separated with an Applied Biosystems 3500XL Genetic Analyser. Developmental validation studies indicated that the 19-plex typing system was accurate, reliable and sensitive, which could also deconvolute mixed C. sativa samples. Specifically, the sensitivity study showed that a full genotyping profile was obtainable with as low as 125 pg of C. sativa DNA. The species specificity study demonstrated that this multiplex has no cross-reactivity with common non C. sativa DNA. In the population study, a total of 162 alleles at 15 autosomal STRs and 14 alleles at two X-chromosome STRs were detected among 85 samples. The efficiency parameters, including the total discrimination power (TDP) and the combined power of exclusion (CPE) of the system, were calculated to exceed 0.999 999 999 999 988 and 0.998 455 889 684 078, respectively, further proving that the system could meet the needs of individual identification. To the extent of the known studies, this is the first study that included the C. sativa sex-determining marker. In conclusion, the developed new 19-plex STR typing system can successfully achieve the purposes of species identification, gender determination, and individual identification, which could be a powerful tool in tracing trade routes of particular drug syndicates or dealers or in linking certain C. sativa to a crime scene.
Keywords: Cannabis sativa; capillary electrophoresis (CE); developmental validation; multiplex system; polymerase chain reaction (PCR); short tandem repeats (STRs).
Copyright © 2022 Xia, Tao, Qu, Zhang, Yu, Yuan, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


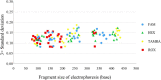
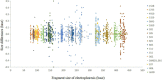
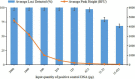
References
-
- Chakraborty R., Stivers D. N., Su B., Zhong Y., Budowle B. (1999). The utility of short tandem repeat loci beyond human identification: implications for development of new DNA typing systems. Electrophoresis 20 1682–1696. 10.1002/(SICI)1522-2683(19990101)20:8<1682::AID-ELPS1682gt;3.0.CO;2-Z - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

