Role of the environment in transmission of Gram-negative bacteria in two consecutive outbreaks in a haematology-oncology department
- PMID: 35295671
- PMCID: PMC8918851
- DOI: 10.1016/j.infpip.2022.100209
Role of the environment in transmission of Gram-negative bacteria in two consecutive outbreaks in a haematology-oncology department
Abstract
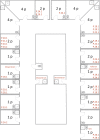
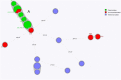
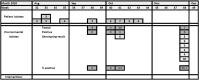
In 2019-2020, two subsequent outbreaks caused by phenotypically identical ESBL-producing Enterobacter cloacae and multi-drug-resistant (MDR) Pseudomonas putida were detected in respectively 15 and 9 patients of the haematology-oncology department. Both bacterial species were resistant to piperacillin-tazobactam, used empirically in (neutropenic) sepsis in our hospital, and ciprofloxacin, used prophylactically in selective digestive decontamination for haematology patients. The E. cloacae outbreak was identified in clinical cultures of blood and urine. Despite intensified infection control measures, new cases were found in weekly point-prevalence screening cultures. Environmental samples of sinks and shower drains appeared positive in 18.1%. To diminish the environmental contamination burden, all siphons of sinks were replaced, and disinfection of sinks and shower drains was intensified using chlorine and soda on a daily basis. Replacement of shower drains was not possible. The outbreak of P. putida remained limited to rectal cultures only, and disappeared spontaneously without interventions. During both outbreaks, multiple strains of the incriminated bacterium were found simultaneously (demonstrated by Amplified-Fragment Length Polymorphism and/or Whole-Genome Multi-locus Sequencing Typing) in patients as well as the environment. It was experimentally shown that a biofilm on the toilet edge may act as a source for nosocomial transmission of Gram-negative bacteria. In conclusion, the drainage system of the hospital is an important reservoir of MDR bacteria, threatening the admitted patients. In existing hospitals, biofilms in the drainage systems cannot be removed. Therefore, it is important that in (re)building plans for hospitals a plan for prevention of nosocomial transmission from environment to patients is incorporated.
Keywords: Gram-negative bacteria; Nosocomial infection; Water points.
© 2022 The Authors.
Figures


References
-
- Kanamori H., Weber D.J., Rutala W.A. Healthcare outbreaks associated with a water reservoir and infection prevention strategies. Clin Infect Dis. 2016;62:1423–1435. - PubMed
-
- Parkes L.O., Hota S.S. Sink-related outbreaks and mitigation strategies in healthcare facilities. Curr Infect Dis Rep. 2018;20:42. - PubMed
-
- Sib E., Voigt A.M., Wilbring G., Schreiber C., Faerber H.A., Skutlarek D., et al. Antibiotic resistant bacteria and resistance genes in biofilms in clinical wastewater networks. Int J Hyg Environ Health. 2019;222:655–662. - PubMed
LinkOut - more resources
Full Text Sources

