Molecular Signatures Correlated With Poor IVF Outcomes: Insights From the mRNA and lncRNA Expression of Endometriotic Granulosa Cells
- PMID: 35295989
- PMCID: PMC8919698
- DOI: 10.3389/fendo.2022.825934
Molecular Signatures Correlated With Poor IVF Outcomes: Insights From the mRNA and lncRNA Expression of Endometriotic Granulosa Cells
Abstract
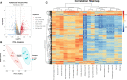
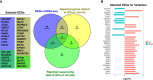
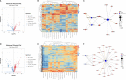
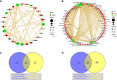
The outcomes of in vitro fertilization (IVF) for endometriotic women are significantly worse than for patients without ovarian endometriosis (OEM), as shown by fewer retrieved oocytes. However, the exact pathophysiological mechanism is still unknown. Thus, we conducted a prospective study that analyzed mRNA and lncRNA transcriptome between granulosa cells (GCs) from patients with fewer retrieved oocytes due to OEM and GCs from controls with male factor (MF) infertility using an RNA sequencing approach. We found a group of significantly differentially expressed genes (DEGs), including NR5A2, MAP3K5, PGRMC2, PRKAR2A, DEPTOR, ITGAV, KPNB1, GPC6, EIF3A, and SMC5, which were validated to be upregulated and negatively correlated with retrieved oocyte numbers in GCs of patients with OEM, while DUSP1 demonstrated the opposite. The molecular functions of these DEGs were mainly enriched in pathways involving mitogen-activated protein kinase (MAPK) signaling, Wnt signaling, steroid hormone response, apoptosis, and cell junction. Furthermore, we performed lncRNA analysis and identified a group of differentially expressed known/novel lncRNAs that were co-expressed with the validated DEGs and correlated with retrieved oocyte numbers. Co-expression networks were constructed between the DEGs and known/novel lncRNAs. These distinctive molecular signatures uncovered in this study are involved in the pathological regulation of ovarian reserve dysfunction in OEM patients.
Keywords: RNA sequencing; female infertility; granulosa cells; lncRNA; mRNA; ovarian endometriosis; retrieved oocyte numbers.
Copyright © 2022 Shi, Wei, Wu, Yuan, Li, Dai, Chen, Zhou, Lin and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Upregulated let-7 expression in the follicular fluid of patients with endometriomas leads to dysfunction of granulosa cells through targeting of IGF1R.Hum Reprod. 2025 Jan 1;40(1):119-137. doi: 10.1093/humrep/deae247. Hum Reprod. 2025. PMID: 39521729 Free PMC article.
-
Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway.Mol Hum Reprod. 2019 Jan 1;25(1):17-29. doi: 10.1093/molehr/gay045. Mol Hum Reprod. 2019. PMID: 30371869
-
Construction of Competing Endogenous RNA Networks Incorporating Transcription Factors to Reveal Differences in Granulosa Cells from Patients with Endometriosis.Genet Test Mol Biomarkers. 2021 Jul;25(7):453-462. doi: 10.1089/gtmb.2020.0152. Genet Test Mol Biomarkers. 2021. PMID: 34280006
-
Novel therapeutic targets to improve IVF outcomes in endometriosis patients: a review and future prospects.Hum Reprod Update. 2021 Aug 20;27(5):923-972. doi: 10.1093/humupd/dmab014. Hum Reprod Update. 2021. PMID: 33930149 Review.
-
Inside the granulosa transcriptome.Gynecol Endocrinol. 2016 Dec;32(12):951-956. doi: 10.1080/09513590.2016.1223288. Epub 2016 Oct 18. Gynecol Endocrinol. 2016. PMID: 27750442 Review.
Cited by
-
Upregulated let-7 expression in the follicular fluid of patients with endometriomas leads to dysfunction of granulosa cells through targeting of IGF1R.Hum Reprod. 2025 Jan 1;40(1):119-137. doi: 10.1093/humrep/deae247. Hum Reprod. 2025. PMID: 39521729 Free PMC article.
-
Impacts of endometrioma on ovarian aging from basic science to clinical management.Front Endocrinol (Lausanne). 2023 Jan 4;13:1073261. doi: 10.3389/fendo.2022.1073261. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36686440 Free PMC article. Review.
-
Decreased oocyte quality in patients with endometriosis is closely related to abnormal granulosa cells.Front Endocrinol (Lausanne). 2023 Aug 16;14:1226687. doi: 10.3389/fendo.2023.1226687. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37664845 Free PMC article. Review.
-
Multiomic insight into the involvement of cell aging related genes in the pathogenesis of endometriosis.Sci Rep. 2025 Apr 23;15(1):14103. doi: 10.1038/s41598-025-96711-2. Sci Rep. 2025. PMID: 40269081 Free PMC article.
-
Sterigmatocystin declines mouse oocyte quality by inducing ferroptosis and asymmetric division defects.J Ovarian Res. 2024 Aug 28;17(1):175. doi: 10.1186/s13048-024-01499-w. J Ovarian Res. 2024. PMID: 39198920 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

