LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer
- PMID: 35296335
- PMCID: PMC8925194
- DOI: 10.1186/s12943-022-01557-1
LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer
Abstract
Background: Histone lysine-specific demethylase 1 (LSD1) expression has been shown to be significantly elevated in gastric cancer (GC) and may be associated with the proliferation and metastasis of GC. It has been reported that LSD1 repressed tumor immunity through programmed cell death 1 ligand 1 (PD-L1) in melanoma and breast cancer. The role of LSD1 in the immune microenvironment of GC is unknown.
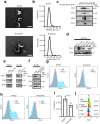
Methods: Expression LSD1 and PD-L1 in GC patients was analyzed by immunohistochemical (IHC) and Western blotting. Exosomes were isolated from the culture medium of GC cells using an ultracentrifugation method and characterized by transmission electronic microscopy (TEM), nanoparticle tracking analysis (NTA), sucrose gradient centrifugation, and Western blotting. The role of exosomal PD-L1 in T-cell dysfunction was assessed by flow cytometry, T-cell killing and enzyme-linked immunosorbent assay (ELISA).
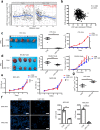
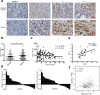
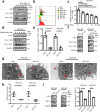
Results: Through in vivo exploration, mouse forestomach carcinoma (MFC) cells with LSD1 knockout (KO) showed significantly slow growth in 615 mice than T-cell-deficient BALB/c nude mice. Meanwhile, in GC specimens, expression of LSD1 was negatively correlated with that of CD8 and positively correlated with that of PD-L1. Further study showed that LSD1 inhibited the response of T cells in the microenvironment of GC by inducing the accumulation of PD-L1 in exosomes, while the membrane PD-L1 stayed constant in GC cells. Using exosomes as vehicles, LSD1 also obstructed T-cell response of other cancer cells while LSD1 deletion rescued T-cell function. It was found that while relying on the existence of LSD1 in donor cells, exosomes can regulate MFC cells proliferation with distinct roles depending on exosomal PD-L1-mediated T-cell immunity in vivo.
Conclusion: LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in GC; this finding indicates a new mechanism with which LSD1 may regulate cancer immunity in GC and provides a new target for immunotherapy against GC.
Keywords: Exosomes; Gastric cancer; LSD1; PD-L1; Tumor immunity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


References
-
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. - PubMed
-
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. - PubMed
-
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. - PubMed
-
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. - PubMed
-
- Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum Mol Genet. 2016;25:2578–2587. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

