Integrative analysis reveals the functional implications and clinical relevance of pyroptosis in low-grade glioma
- PMID: 35296768
- PMCID: PMC8925295
- DOI: 10.1038/s41598-022-08619-w
Integrative analysis reveals the functional implications and clinical relevance of pyroptosis in low-grade glioma
Abstract
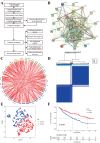
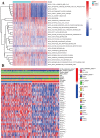
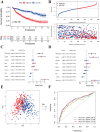
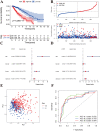
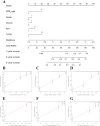
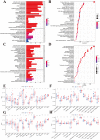
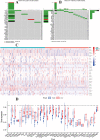
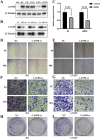
Using the Chinese Glioma Genome Atlas (training dataset) and The Cancer Genome Atlas (validation dataset), we found that low-grade gliomas can be divided into two molecular subclasses based on 30 pyroptosis genes. Cluster 1 presented higher immune cell and immune function scores and poorer prognosis than Cluster 2. We established a prognostic model based on 10 pyroptosis genes; the model could predict overall survival in glioma and was well validated in an independent dataset. The high-risk group had relatively higher immune cell and immune function scores and lower DNA methylation levels in pyroptosis genes than the low-risk group. There were no marked differences in pyroptosis gene alterations between the high- and low-risk groups. The competing endogenous RNA (ceRNA) regulatory network uncovered the lncRNA-miRNA-mRNA regulation patterns of the different risk groups in low-grade glioma. Five pairs of target genes and drugs were identified. In vitro, CASP8 silencing inhibited the migration and invasion of glioma cells. The expression of pyroptosis genes can reflect the molecular biological and clinical features of low-grade glioma subclasses. The developed prognostic model can predict overall survival and distinguish molecular alterations in patients. Our integrated analyses could provide valuable guidelines for improving risk management and therapy for low-grade glioma patients.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

