Human gut bacteria produce ΤΗ17-modulating bile acid metabolites
- PMID: 35296854
- PMCID: PMC9132548
- DOI: 10.1038/s41586-022-04480-z
Human gut bacteria produce ΤΗ17-modulating bile acid metabolites
Abstract
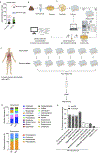
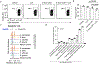
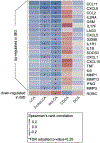
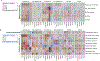
The microbiota modulates gut immune homeostasis. Bacteria influence the development and function of host immune cells, including T helper cells expressing interleukin-17A (TH17 cells). We previously reported that the bile acid metabolite 3-oxolithocholic acid (3-oxoLCA) inhibits TH17 cell differentiation1. Although it was suggested that gut-residing bacteria produce 3-oxoLCA, the identity of such bacteria was unknown, and it was unclear whether 3-oxoLCA and other immunomodulatory bile acids are associated with inflammatory pathologies in humans. Here we identify human gut bacteria and corresponding enzymes that convert the secondary bile acid lithocholic acid into 3-oxoLCA as well as the abundant gut metabolite isolithocholic acid (isoLCA). Similar to 3-oxoLCA, isoLCA suppressed TH17 cell differentiation by inhibiting retinoic acid receptor-related orphan nuclear receptor-γt, a key TH17-cell-promoting transcription factor. The levels of both 3-oxoLCA and isoLCA and the 3α-hydroxysteroid dehydrogenase genes that are required for their biosynthesis were significantly reduced in patients with inflammatory bowel disease. Moreover, the levels of these bile acids were inversely correlated with the expression of TH17-cell-associated genes. Overall, our data suggest that bacterially produced bile acids inhibit TH17 cell function, an activity that may be relevant to the pathophysiology of inflammatory disorders such as inflammatory bowel disease.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
A.S.D. is a consultant for Takeda Pharmaceuticals and Axial Therapeutics. J.R.H. is a consultant for CJ Research Center, LLC and Interon Laboratories and on the scientific advisory board for ChunLab. P.J.T. is on the scientific advisory board for Kaleido, Pendulum, Seres, and SNIPRbiome. C.H. is on the scientific advisory boards of Seres Therapeutics, Empress Therapeutics, and ZOE Nutrition.
Figures






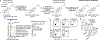

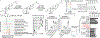
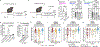
Comment in
-
Expanding Links Between Gut Microbiota and Bowel Inflammation.Gastroenterology. 2022 Jul;163(1):333-334. doi: 10.1053/j.gastro.2022.05.001. Epub 2022 May 4. Gastroenterology. 2022. PMID: 35525319 No abstract available.
References
-
- Fiorucci S & Distrutti E Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med 21, 702–714 (2015). - PubMed
-
- Ridlon JM, Kang D-J & Hylemon PB Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47, 241–259 (2005). - PubMed
-
- Schaap FG, Trauner M & Jansen PLM Bile acid receptors as targets for drug development. Nat Rev Gastroentero 11, 55–67 (2014). - PubMed
MeSH terms
Substances
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R00 AI147165/AI/NIAID NIH HHS/United States
- R01 DK110559/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 DK120985/DK/NIDDK NIH HHS/United States
- R35 GM128618/GM/NIGMS NIH HHS/United States
- K99 AI147165/AI/NIAID NIH HHS/United States
- T32 GM095450/GM/NIGMS NIH HHS/United States
- R24 DK110499/DK/NIDDK NIH HHS/United States
- 210664/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- U54 DE023798/DE/NIDCR NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- R01 DK114252/DK/NIDDK NIH HHS/United States
- R01 AR074500/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

