A genome-scale screen for synthetic drivers of T cell proliferation
- PMID: 35296855
- PMCID: PMC9908437
- DOI: 10.1038/s41586-022-04494-7
A genome-scale screen for synthetic drivers of T cell proliferation
Abstract
The engineering of autologous patient T cells for adoptive cell therapies has revolutionized the treatment of several types of cancer1. However, further improvements are needed to increase response and cure rates. CRISPR-based loss-of-function screens have been limited to negative regulators of T cell functions2-4 and raise safety concerns owing to the permanent modification of the genome. Here we identify positive regulators of T cell functions through overexpression of around 12,000 barcoded human open reading frames (ORFs). The top-ranked genes increased the proliferation and activation of primary human CD4+ and CD8+ T cells and their secretion of key cytokines such as interleukin-2 and interferon-γ. In addition, we developed the single-cell genomics method OverCITE-seq for high-throughput quantification of the transcriptome and surface antigens in ORF-engineered T cells. The top-ranked ORF-lymphotoxin-β receptor (LTBR)-is typically expressed in myeloid cells but absent in lymphocytes. When overexpressed in T cells, LTBR induced profound transcriptional and epigenomic remodelling, leading to increased T cell effector functions and resistance to exhaustion in chronic stimulation settings through constitutive activation of the canonical NF-κB pathway. LTBR and other highly ranked genes improved the antigen-specific responses of chimeric antigen receptor T cells and γδ T cells, highlighting their potential for future cancer-agnostic therapies5. Our results provide several strategies for improving next-generation T cell therapies by the induction of synthetic cell programmes.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures









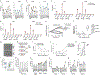




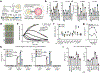
References
Extended Data References
-
- Ding J et al. Systematic comparative analysis of single cell RNA-sequencing methods. http://biorxiv.org/lookup/doi/10.1101/632216 (2019) doi:10.1101/632216. - DOI - DOI
References
-
- Abramson JS et al. Transcend NHL 001: Immunotherapy with the CD19-Directed CAR T-Cell Product JCAR017 Results in High Complete Response Rates in Relapsed or Refractory B-Cell Non-Hodgkin Lymphoma. Blood 128, 4192–4192 (2016).
Additional References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

