Immune regulation by fungal strain diversity in inflammatory bowel disease
- PMID: 35296857
- PMCID: PMC9166917
- DOI: 10.1038/s41586-022-04502-w
Immune regulation by fungal strain diversity in inflammatory bowel disease
Erratum in
-
Author Correction: Immune regulation by fungal strain diversity in inflammatory bowel disease.Nature. 2022 Aug;608(7922):E21. doi: 10.1038/s41586-022-05102-4. Nature. 2022. PMID: 35859182 No abstract available.
Abstract
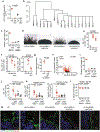
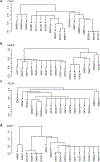
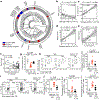
The fungal microbiota (mycobiota) is an integral part of the complex multikingdom microbial community colonizing the mammalian gastrointestinal tract and has an important role in immune regulation1-6. Although aberrant changes in the mycobiota have been linked to several diseases, including inflammatory bowel disease3-9, it is currently unknown whether fungal species captured by deep sequencing represent living organisms and whether specific fungi have functional consequences for disease development in affected individuals. Here we developed a translational platform for the functional analysis of the mycobiome at the fungal-strain- and patient-specific level. Combining high-resolution mycobiota sequencing, fungal culturomics and genomics, a CRISPR-Cas9-based fungal strain editing system, in vitro functional immunoreactivity assays and in vivo models, this platform enables the examination of host-fungal crosstalk in the human gut. We discovered a rich genetic diversity of opportunistic Candida albicans strains that dominate the colonic mucosa of patients with inflammatory bowel disease. Among these human-gut-derived isolates, strains with high immune-cell-damaging capacity (HD strains) reflect the disease features of individual patients with ulcerative colitis and aggravated intestinal inflammation in vivo through IL-1β-dependent mechanisms. Niche-specific inflammatory immunity and interleukin-17A-producing T helper cell (TH17 cell) antifungal responses by HD strains in the gut were dependent on the C. albicans-secreted peptide toxin candidalysin during the transition from a benign commensal to a pathobiont state. These findings reveal the strain-specific nature of host-fungal interactions in the human gut and highlight new diagnostic and therapeutic targets for diseases of inflammatory origin.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
Cornell University has filed a provisional patent application covering inventions described in this manuscript. The authors declare no other competing interests.
Figures











Comment in
-
Fungal strain-dependent inflammation in IBD.Nat Rev Gastroenterol Hepatol. 2022 May;19(5):280. doi: 10.1038/s41575-022-00613-x. Nat Rev Gastroenterol Hepatol. 2022. PMID: 35379940 No abstract available.
-
Expanding Links Between Gut Microbiota and Bowel Inflammation.Gastroenterology. 2022 Jul;163(1):333-334. doi: 10.1053/j.gastro.2022.05.001. Epub 2022 May 4. Gastroenterology. 2022. PMID: 35525319 No abstract available.
-
Paying attention to minutiae: Strain level differences drive disease etiology.Med. 2022 May 13;3(5):270-272. doi: 10.1016/j.medj.2022.04.011. Epub 2022 May 13. Med. 2022. PMID: 35584642
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

