Sindbis Macrodomain Poly-ADP-Ribose Hydrolase Activity Is Important for Viral RNA Synthesis
- PMID: 35297669
- PMCID: PMC9006893
- DOI: 10.1128/jvi.01516-21
Sindbis Macrodomain Poly-ADP-Ribose Hydrolase Activity Is Important for Viral RNA Synthesis
Abstract
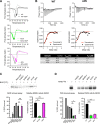
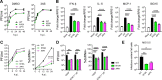
ADP-ribosylation is a highly dynamic posttranslational modification frequently studied in stress response pathways with recent attention given to its role in response to viral infection. Notably, the alphaviruses encode catalytically active macrodomains capable of ADP-ribosylhydrolase (ARH) activities, implying a role in remodeling the cellular ADP-ribosylome. This report decouples mono- and poly-ARH contributions to macrodomain function using a newly engineered Sindbis virus (SINV) mutant with attenuated poly-ARH activity. Our findings indicate that viral poly-ARH activity is uniquely required for high titer replication in mammalian systems. Despite translating incoming genomic RNA as efficiently as WT virus, mutant viruses have a reduced capacity to establish productive infection, offering a more complete understanding of the kinetics and role of the alphavirus macrodomain with important implications for broader ADP-ribosyltransferase biology. IMPORTANCE Viral macrodomains have drawn attention in recent years due to their high degree of conservation in several virus families (e.g., coronaviruses and alphaviruses) and their potential druggability. These domains erase mono- or poly-ADP-ribose, posttranslational modifications written by host poly-ADP-ribose polymerase (PARP) proteins, from undetermined host or viral proteins to enhance replication. Prior work determined that efficient alphavirus replication requires catalytically active macrodomains; however, which form of the modification requires removal and from which protein(s) had not been determined. Here, we present evidence for the specific requirement of poly-ARH activity to ensure efficient productive infection and virus replication.
Keywords: ADP-ribosylation; PARP/ARTD; RNA synthesis; Sindbis virus; alphavirus; virus replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

