The Human Liver-Expressed Lectin CD302 Restricts Hepatitis C Virus Infection
- PMID: 35297672
- PMCID: PMC9006913
- DOI: 10.1128/jvi.01995-21
The Human Liver-Expressed Lectin CD302 Restricts Hepatitis C Virus Infection
Abstract
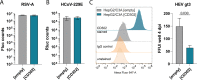
C-type lectin domain-containing proteins (CTLDcps) shape host responses to pathogens and infectious disease outcomes. Previously, we identified the murine CTLDcp Cd302 as restriction factor, limiting hepatitis C virus (HCV) infection of murine hepatocytes. In this study, we investigated in detail the human orthologue's ability to restrict HCV infection in human liver cells. CD302 overexpression in Huh-7.5 cells potently inhibited infection of diverse HCV chimeras representing seven genotypes. Transcriptional profiling revealed abundant CD302 mRNA expression in human hepatocytes, the natural cellular target of HCV. Knockdown of endogenously expressed CD302 modestly enhanced HCV infection of Huh-7.5 cells and primary human hepatocytes. Functional analysis of naturally occurring CD302 transcript variants and engineered CD302 mutants showed that the C-type lectin-like domain (CTLD) is essential for HCV restriction, whereas the cytoplasmic domain (CPD) is dispensable. Coding single nucleotide polymorphisms occurring in human populations and mapping to different domains of CD302 did not influence the capacity of CD302 to restrict HCV. Assessment of the anti-HCV phenotype at different life cycle stages indicated that CD302 preferentially targets the viral entry step. In contrast to the murine orthologue, overexpression of human CD302 did not modulate downstream expression of nuclear receptor-controlled genes. Ectopic CD302 expression restricted infection of liver tropic hepatitis E virus (HEV), while it did not affect infection rates of two respiratory viruses, including respiratory syncytial virus (RSV) and the alpha coronavirus HVCoV-229E. Together, these findings suggest that CD302 contributes to liver cell-intrinsic defense against HCV and might mediate broader antiviral defenses against additional hepatotropic viruses. IMPORTANCE The liver represents an immunoprivileged organ characterized by enhanced resistance to immune responses. However, the importance of liver cell-endogenous, noncytolytic innate immune responses in pathogen control is not well defined. Although the role of myeloid cell-expressed CTLDcps in host responses to viruses has been characterized in detail, we have little information about their potential functions in the liver and their relevance for immune responses in this organ. Human hepatocytes endogenously express the CTLDcp CD302. Here, we provide evidence that CD302 limits HCV infection of human liver cells, likely by inhibiting a viral cell entry step. We confirm that the dominant liver-expressed transcript variant, as well as naturally occurring coding variants of CD302, maintain the capacity to restrict HCV. We further show that the CTLD of the protein is critical for the anti-HCV activity and that overexpressed CD302 limits HEV infection. Thus, CD302 likely contributes to human liver-intrinsic antiviral defenses.
Keywords: hepatitis C virus; hepatitis E virus; lectin; mechanism; polymorphism; transcript variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- World Health Organization. 2017. Global viral hepatitis report, 2017. World Health Organization, Geneva, Switzerland.
-
- Midgard H, Bjøro B, Mæland A, Konopski Z, Kileng H, Damås JK, Paulsen J, Heggelund L, Sandvei PK, Ringstad JO, Karlsen LN, Stene-Johansen K, Pettersson JH-O, Dorenberg DH, Dalgard O. 2016. Hepatitis C reinfection after sustained virological response. J Hepatol 64:1020–1026. 10.1016/j.jhep.2016.01.001. - DOI - PubMed
-
- Pineda JA, Nunez-Torres R, Tellez F, Mancebo M, Garcia F, Merchante N, Perez-Perez M, Neukam K, Macias J, Real LM, HEPAVIR Group of The Andalusian Society of Infectious Diseases (SAEI). 2015. Hepatitis C virus reinfection after sustained virological response in HIV-infected patients with chronic hepatitis C. J Infect 71:571–577. 10.1016/j.jinf.2015.07.006. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

