A Polymorphic Gene within the Mycobacterium smegmatis esx1 Locus Determines Mycobacterial Self-Identity and Conjugal Compatibility
- PMID: 35297678
- PMCID: PMC9040860
- DOI: 10.1128/mbio.00213-22
A Polymorphic Gene within the Mycobacterium smegmatis esx1 Locus Determines Mycobacterial Self-Identity and Conjugal Compatibility
Abstract
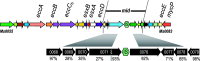
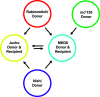
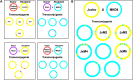
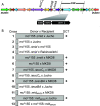
Mycobacteria mediate horizontal gene transfer (HGT) by a process called distributive conjugal transfer (DCT) that is mechanistically distinct from oriT-mediated plasmid transfer. The transfer of multiple, independent donor chromosome segments generates transconjugants with genomes that are mosaic blends of their parents. Previously, we had characterized contact-dependent conjugation between two independent isolates of Mycobacterium smegmatis. Here, we expand our analyses to include five independent isolates of M. smegmatis and establish that DCT is both active and prevalent among natural isolates of M. smegmatis. Two of these five strains were recipients but exhibited distinct conjugal compatibilities with donor strains, suggesting an ability to distinguish between potential donor partners. We determined that a single gene, Msmeg0070, was responsible for conferring mating compatibility using a combination of comparative DNA sequence analysis, bacterial genome-wide association studies (GWAS), and targeted mutagenesis. Msmeg0070 maps within the esx1 secretion locus, and we establish that it confers mycobacterial self-identity with parallels to kin recognition. Similar to other kin model systems, orthologs of Msmeg0070 are highly polymorphic. The identification of a kin recognition system in M. smegmatis reinforces the concept that communication between cells is an important checkpoint prior to DCT commitment and implies that there are likely to be other, unanticipated forms of social behaviors in mycobacteria. IMPORTANCE Conjugation, unlike other forms of HGT, requires direct interaction between two viable bacteria, which must be capable of distinguishing between mating types to allow successful DNA transfer from donor to recipient. We show that the conjugal compatibility of Mycobacterium smegmatis isolates is determined by a single, polymorphic gene located within the conserved esx1 secretion locus. This gene confers self-identity; the expression of identical Msmeg0070 proteins in both donor-recipient partners prevents DNA transfer. The presence of this polymorphic locus in many environmental mycobacteria suggests that kin identification is important in promoting beneficial gene flow between nonkin mycobacteria. Cell-cell communication, mediated by kin recognition and ESX secretion, is a key checkpoint in mycobacterial conjugation and likely plays a more global role in mycobacterial biology.
Keywords: conjugation; esx1; horizontal gene transfer; kin recognition; mycobacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

