Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection
- PMID: 35297757
- PMCID: PMC9176291
- DOI: 10.1099/mgen.0.000751
Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection
Abstract
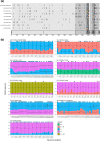
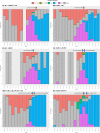
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has infected almost 200 million people worldwide by July 2021 and the pandemic has been characterized by infection waves of viral lineages showing distinct fitness profiles. The simultaneous infection of a single individual by two distinct SARS-CoV-2 lineages may impact COVID-19 disease progression and provides a window of opportunity for viral recombination and the emergence of new lineages with differential phenotype. Several hundred SARS-CoV-2 lineages are currently well phylogenetically defined, but two main factors have precluded major coinfection/codetection and recombination analysis thus far: (i) the low diversity of SARS-CoV-2 lineages during the first year of the pandemic, which limited the identification of lineage defining mutations necessary to distinguish coinfecting/recombining viral lineages; and the (ii) limited availability of raw sequencing data where abundance and distribution of intrasample/intrahost variability can be accessed. Here, we assembled a large sequencing dataset from Brazilian samples covering a period of 18 May 2020 to 30 April 2021 and probed it for unexpected patterns of high intrasample/intrahost variability. This approach enabled us to detect nine cases of SARS-CoV-2 coinfection with well characterized lineage-defining mutations, representing 0.61 % of all samples investigated. In addition, we matched these SARS-CoV-2 coinfections with spatio-temporal epidemiological data confirming its plausibility with the cocirculating lineages at the timeframe investigated. Our data suggests that coinfection with distinct SARS-CoV-2 lineages is a rare phenomenon, although it is certainly a lower bound estimate considering the difficulty to detect coinfections with very similar SARS-CoV-2 lineages and the low number of samples sequenced from the total number of infections.
Keywords: COVID-19; codetection; coinfection; genomics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Mullen JL, Tsueng G, Abdel Latif A, Alkuzweny M, Cano M, et al. outbreak.info. 2021. https://outbreak.info/
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

