Regenerating vascular mural cells in zebrafish fin blood vessels are not derived from pre-existing mural cells and differentially require Pdgfrb signalling for their development
- PMID: 35297968
- PMCID: PMC9058498
- DOI: 10.1242/dev.199640
Regenerating vascular mural cells in zebrafish fin blood vessels are not derived from pre-existing mural cells and differentially require Pdgfrb signalling for their development
Abstract
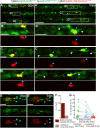
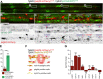
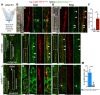
Vascular networks comprise endothelial cells and mural cells, which include pericytes and smooth muscle cells. To elucidate the mechanisms controlling mural cell recruitment during development and tissue regeneration, we studied zebrafish caudal fin arteries. Mural cells colonizing arteries proximal to the body wrapped around them, whereas those in more distal regions extended protrusions along the proximo-distal vascular axis. Both cell populations expressed platelet-derived growth factor receptor β (pdgfrb) and the smooth muscle cell marker myosin heavy chain 11a (myh11a). Most wrapping cells in proximal locations additionally expressed actin alpha2, smooth muscle (acta2). Loss of Pdgfrb signalling specifically decreased mural cell numbers at the vascular front. Using lineage tracing, we demonstrate that precursor cells located in periarterial regions and expressing Pgdfrb can give rise to mural cells. Studying tissue regeneration, we did not find evidence that newly formed mural cells were derived from pre-existing cells. Together, our findings reveal conserved roles for Pdgfrb signalling in development and regeneration, and suggest a limited capacity of mural cells to self-renew or contribute to other cell types during tissue regeneration.
Keywords: Blood vessel; Caudal fin; Mural cell; Pdgfrb signalling; Tissue regeneration; Zebrafish.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures






References
-
- Ando, K., Shih, Y.-H., Ebarasi, L., Grosse, A., Portman, D., Chiba, A., Mattonet, K., Gerri, C., Stainier, D. Y. R., Mochizuki, N.et al. (2021). Conserved and context-dependent roles for pdgfrb signaling during zebrafish vascular mural cell development. Dev. Biol. 479, 11-22. 10.1016/j.ydbio.2021.06.010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

