MicroRNA Dysregulation in Prostate Cancer
- PMID: 35300057
- PMCID: PMC8923686
- DOI: 10.2147/PGPM.S348565
MicroRNA Dysregulation in Prostate Cancer
Abstract
Prostate cancer biology is complex, and needs to be deciphered. The latest evidence reveals the significant role of non-coding RNAs, particularly microRNAs (miRNAs), as key regulatory factors in cancer. Therefore, the identification of altered miRNA patterns involved in prostate cancer will allow them to be used for development of novel diagnostic and prognostic biomarkers.
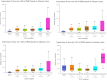
Patients and methods: We performed a miRNAs transcriptomic analysis, using microarray (10 matched pairs tumor tissue versus normal adjacent tissue, selected based on inclusion criteria), followed by overlapping with TCGA data. A total of 292 miRNAs were differentially expressed, with 125 upregulated and 167 downregulated in TCGA patients' cohort with PRAD (prostate adenocarcinoma), respectively for the microarray experiments; 16 upregulated and 44 downregulated miRNAs were found in our cohort. To confirm our results obtained for tumor tissue, we performed validation with qRT-PCR at the tissue and plasma level of two selected transcripts, and finally, we focused on the identification of altered miRNAs involved in key biological processes.
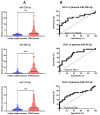
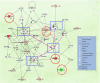
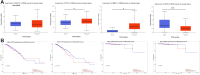
Results: A common signature identified a panel of 12 upregulated and 1 downregulated miRNA, targeting and interconnected in a network with the TP53, AGO2, BIRC5 gene and EGFR as a core element. Among this signature, the overexpressed transcripts (miR-20b-5p, miR-96-5p, miR-183-5p) and the downregulated miR-542-5p were validated by qRT-PCR in an additional patients' cohort of 34 matched tumor and normal adjacent paired samples. Further, we performed the validation of the expression level for miR-20b-5p, miR-96-5p, miR-183-5p plasma, on the same patients' cohort versus a healthy control group, confirming the overexpression of these transcripts in the PRAD group, demonstrating the liquid biopsy as a potential investigational tool in prostate cancer.
Conclusion: In this pilot study, we provide evidence on miRNA dysregulation and its association with key functional components of the PRAD landscape, where an important role is acted by miR-20b-5p, miR-542-5p, or the oncogenic cluster miR-183-96-182.
Keywords: biological network; microRNA; molecular signature; plasma; prostate adenocarcinoma; tissue.
© 2022 Schitcu et al.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

