Short open reading frames (sORFs) and microproteins: an update on their identification and validation measures
- PMID: 35300685
- PMCID: PMC8928697
- DOI: 10.1186/s12929-022-00802-5
Short open reading frames (sORFs) and microproteins: an update on their identification and validation measures
Abstract
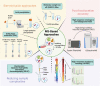
A short open reading frame (sORFs) constitutes ≤ 300 bases, encoding a microprotein or sORF-encoded protein (SEP) which comprises ≤ 100 amino acids. Traditionally dismissed by genome annotation pipelines as meaningless noise, sORFs were found to possess coding potential with ribosome profiling (RIBO-Seq), which unveiled sORF-based transcripts at various genome locations. Nonetheless, the existence of corresponding microproteins that are stable and functional was little substantiated by experimental evidence initially. With recent advancements in multi-omics, the identification, validation, and functional characterisation of sORFs and microproteins have become feasible. In this review, we discuss the history and development of an emerging research field of sORFs and microproteins. In particular, we focus on an array of bioinformatics and OMICS approaches used for predicting, sequencing, validating, and characterizing these recently discovered entities. These strategies include RIBO-Seq which detects sORF transcripts via ribosome footprints, and mass spectrometry (MS)-based proteomics for sequencing the resultant microproteins. Subsequently, our discussion extends to the functional characterisation of microproteins by incorporating CRISPR/Cas9 screen and protein-protein interaction (PPI) studies. Our review discusses not only detection methodologies, but we also highlight on the challenges and potential solutions in identifying and validating sORFs and their microproteins. The novelty of this review lies within its validation for the functional role of microproteins, which could contribute towards the future landscape of microproteomics.
Keywords: Mass spectrometry; Microproteins; Proteogenomics; Ribosome profiling (RIBO-Seq); Short open reading frame (sORF); Small open reading frame (smORF).
© 2022. The Author(s).
Conflict of interest statement
The authors have declared no conflict of interest.
Figures



References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Gates AJ, Gysi DM, Kellis M, Barabási A-L. A wealth of discovery built on the Human Genome Project—by the numbers. Nature. 2021;590:212–215. - PubMed
-
- Skovgaard M, Jensen LJ, Brunak S, Ussery D, Krogh A. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001 [cited 2021 Apr 15]. p. 425–8. https://linkinghub.elsevier.com/retrieve/pii/S0168952501023721. Accessed 15 Apr 2021. - PubMed
-
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

