Unraveling the role of the microbiome in chronic rhinosinusitis
- PMID: 35300985
- PMCID: PMC9354834
- DOI: 10.1016/j.jaci.2022.02.022
Unraveling the role of the microbiome in chronic rhinosinusitis
Abstract
Chronic rhinosinusitis (CRS) is a complex, heterogenous condition that is likely associated with infectious and inflammatory causative factors. Renewed interest in the role that microbes play in this condition has stemmed from advancements in microbe identification and parallel research implicating the microbiome as having a role in other chronic inflammatory conditions. This clinical commentary provides a review of the current literature relevant to chronic rhinosinusitis. Particular focus is placed on factors specific to investigation of the sinonasal microbiome, evidence for the role of dysbiosis in the disease state, and influences that may affect the microbiome. Possible mechanisms of disease and therapeutic implications through microbial manipulation are also reviewed, as are deficiencies and limitations of the current body of research.
Keywords: Microbiome; chronic rhinosinusitis; host-microbial interactions; microbiome manipulation; next-generation sequencing; probiotic; therapeutic intervention.
Copyright © 2022 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Figures
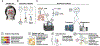
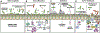
References
-
- Wagner Mackenzie B, Waite DW, Hoggard M, Douglas RG, Taylor MW, Biswas K. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol 2017;19:381–92. - PubMed
-
- Hoggard M, Biswas K, Zoing M, Wagner Mackenzie B, Taylor MW, Douglas RG. Evidence of microbiota dysbiosis in chronic rhinosinusitis. Int Forum Allergy Rhinol 2017;7:230–9. - PubMed
-
- Lal D, Keim P, Delisle J, Barker B, Rank MA, Chia N, et al. Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int Forum Allergy Rhinol 2017;7:561–9. - PubMed

