Multi-omic cross-sectional cohort study of pre-malignant Barrett's esophagus reveals early structural variation and retrotransposon activity
- PMID: 35301290
- PMCID: PMC8931005
- DOI: 10.1038/s41467-022-28237-4
Multi-omic cross-sectional cohort study of pre-malignant Barrett's esophagus reveals early structural variation and retrotransposon activity
Abstract
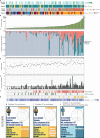
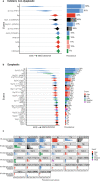
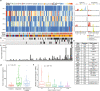
Barrett's esophagus is a pre-malignant lesion that can progress to esophageal adenocarcinoma. We perform a multi-omic analysis of pre-cancer samples from 146 patients with a range of outcomes, comprising 642 person years of follow-up. Whole genome sequencing reveals complex structural variants and LINE-1 retrotransposons, as well as known copy number changes, occurring even prior to dysplasia. The structural variant burden captures the most variance across the cohort and genomic profiles do not always match consensus clinical pathology dysplasia grades. Increasing structural variant burden is associated with: high levels of chromothripsis and breakage-fusion-bridge events; increased expression of genes related to cell cycle checkpoint, DNA repair and chromosomal instability; and epigenetic silencing of Wnt signalling and cell cycle genes. Timing analysis reveals molecular events triggering genomic instability with more clonal expansion in dysplastic samples. Overall genomic complexity occurs early in the Barrett's natural history and may inform the potential for cancer beyond the clinically discernible phenotype.
© 2022. The Author(s).
Conflict of interest statement
R.C.F. and M.O’.D. are named on patents related to Cytosponge and related assays (Cell Sampling Device PCT/GB2010/002077, Biomarker for Barrett’s Oesophagus US2012/0009597) which have been licensed by the Medical Research Council to Covidien GI Solutions (now Medtronic) and are co-founders of CYTED Ltd. R.C.F. and M.O’.D. have a patent on a method for detecting benign conditions in the upper intestinal tract (Methods for Detection of Benign Conditions PCT/GB2012/000855). These are not directly involved in the topic of this paper. The remaining authors declare no competing interests.
Figures





References
-
- Pashayan N, Pharoah PDP. The challenge of early detection in cancer. Science. 2020;368:589–590. - PubMed
-
- Zagari RM, et al. Gastro-oesophageal reflux symptoms, oesophagitis and barrett’s oesophagus in the general population: the Loiano-Monghidoro study. Gut. 2008;57:1354–1359. - PubMed
-
- Ronkainen J, et al. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–1831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

