Novel In Silico mRNA vaccine design exploiting proteins of M. tuberculosis that modulates host immune responses by inducing epigenetic modifications
- PMID: 35301360
- PMCID: PMC8929471
- DOI: 10.1038/s41598-022-08506-4
Novel In Silico mRNA vaccine design exploiting proteins of M. tuberculosis that modulates host immune responses by inducing epigenetic modifications
Abstract
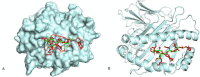
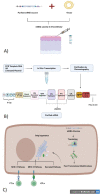
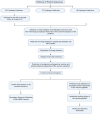
Tuberculosis is an airborne infectious disease caused by Mycobacterium tuberculosis. BCG is the only approved vaccine. However, it has limited global efficacy. Pathogens could affect the transcription of host genes, especially the ones related to the immune system, by inducing epigenetic modifications. Many proteins of M. tuberculosis were found to affect the host's epigenome. Nine proteins were exploited in this study to predict epitopes to develop an mRNA vaccine against tuberculosis. Many immunoinformatics tools were employed to construct this vaccine to elicit cellular and humoral immunity. We performed molecular docking between selected epitopes and their corresponding MHC alleles. Thirty epitopes, an adjuvant TLR4 agonist RpfE, constructs for subcellular trafficking, secretion booster, and specific linkers were combined to develop the vaccine. This proposed construct was tested to cover 99.38% of the population. Moreover, it was tested to be effective and safe. An in silico immune simulation of the vaccine was also performed to validate our hypothesis. It also underwent codon optimization to ensure mRNA's efficient translation once it reaches the cytosol of a human host. Furthermore, secondary and tertiary structures of the vaccine peptide were predicted and docked against TLR-4 and TLR-3.Molecular dynamics simulation was performed to validate the stability of the binding complex. It was found that this proposed construction can be a promising vaccine against tuberculosis. Hence, our proposed construct is ready for wet-lab experiments to approve its efficacy.
© 2022. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures




References
-
- World Health Organization. Global tuberculosis report (2020).
-
- Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. Esx secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016;14:677–691. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

