Common coupling map advances GPCR-G protein selectivity
- PMID: 35302494
- PMCID: PMC9005189
- DOI: 10.7554/eLife.74107
Common coupling map advances GPCR-G protein selectivity
Abstract
Two-thirds of human hormones and one-third of clinical drugs act on membrane receptors that couple to G proteins to achieve appropriate functional responses. While G protein transducers from literature are annotated in the Guide to Pharmacology database, two recent large-scale datasets now expand the receptor-G protein 'couplome'. However, these three datasets differ in scope and reported G protein couplings giving different coverage and conclusions on G protein-coupled receptor (GPCR)-G protein signaling. Here, we report a common coupling map uncovering novel couplings supported by both large-scale studies, the selectivity/promiscuity of GPCRs and G proteins, and how the co-coupling and co-expression of G proteins compare to the families from phylogenetic relationships. The coupling map and insights on GPCR-G protein selectivity will catalyze advances in receptor research and cellular signaling toward the exploitation of G protein signaling bias in design of safer drugs.
Keywords: G protein; GPCR; computational biology; human; pharmacology; signal transduction; systems biology.
© 2022, Hauser et al.
Conflict of interest statement
AH, CA, AI, DG No competing interests declared, CN, AM was an employees of Domain Therapeutics North America during part or all of this research, MB is the president of Domain Therapeutics scientific advisory board
Figures






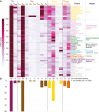

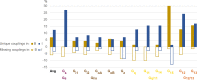
References
-
- Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Southan C, Sharman JL, Campo B, Cavanagh DR, Alexander SPH, Davenport AP, Spedding M, Davies JA, NC-IUPHAR The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Research. 2020;48:D1006–D1021. doi: 10.1093/nar/gkz951. - DOI - PMC - PubMed
-
- Avet C, Mancini A, Breton B, Gouill CL, Hauser AS, Normand C, Kobayashi H, Gross F, Hogue M, Lukasheva V, St-Onge S, Carrier M, Héroux M, Morissette S, Fauman E, Fortin JP, Schann S, Leroy X, Gloriam DE, Bouvier M. Effector membrane translocation biosensors reveal G protein and B-arrestin profiles of 100 therapeutically relevant GPCRs. bioRxiv. 2020 doi: 10.1101/2020.04.20.052027. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

