Spatiotemporally resolved multivariate pattern analysis for M/EEG
- PMID: 35302683
- PMCID: PMC9188977
- DOI: 10.1002/hbm.25835
Spatiotemporally resolved multivariate pattern analysis for M/EEG
Abstract
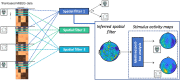
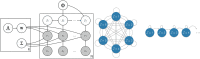
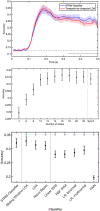
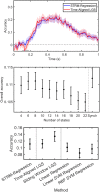
An emerging goal in neuroscience is tracking what information is represented in brain activity over time as a participant completes some task. While electroencephalography (EEG) and magnetoencephalography (MEG) offer millisecond temporal resolution of how activity patterns emerge and evolve, standard decoding methods present significant barriers to interpretability as they obscure the underlying spatial and temporal activity patterns. We instead propose the use of a generative encoding model framework that simultaneously infers the multivariate spatial patterns of activity and the variable timing at which these patterns emerge on individual trials. An encoding model inversion maps from these parameters to the equivalent decoding model, allowing predictions to be made about unseen test data in the same way as in standard decoding methodology. These SpatioTemporally Resolved MVPA (STRM) models can be flexibly applied to a wide variety of experimental paradigms, including classification and regression tasks. We show that these models provide insightful maps of the activity driving predictive accuracy metrics; demonstrate behaviourally meaningful variation in the timing of pattern emergence on individual trials; and achieve predictive accuracies that are either equivalent or surpass those achieved by more widely used methods. This provides a new avenue for investigating the brain's representational dynamics and could ultimately support more flexible experimental designs in the future.
Keywords: EEG; MEG; decoding; encoding; single trial task dynamics.
© 2022 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest..
Figures




References
-
- Beal, M. J. (2003). Variational algorithms for approximate bayesian inference (PhD Thesis [Issue May]). Gatsby Computational Neuroscience Unit, University College London.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

